Formation of Iron Oxide Nanoparticles in the Internal Cavity of Ferritin-Like Dps Protein: Studies by Anomalous X-Ray Scattering
Eleonora V. Shtykova1,a*, Maxim V. Petoukhov1,2, and Andrey A. Mozhaev1,3,4,5
1Shubnikov Institute of Crystallography, Crystallography and Photonics Federal Scientific Research Centre, Russian Academy of Sciences, 119333 Moscow, Russia2Frumkin Institute of Physical Chemistry and Electrochemistry, Russian Academy of Sciences, 119071 Moscow, Russia
3Shemyakin–Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, 117997 Moscow, Russia
4Pirogov Russian National Research Medical University, 117997 Moscow, Russia
5National Research University Higher School of Economics, 101000 Moscow, Russia
* To whom correspondence should be addressed.
Received April 12, 2022; Revised April 26, 2022; Accepted April 26, 2022
DNA-binding protein from starved cells (Dps) takes a special place among dodecamer mini-ferritins. Its most important function is protection of bacterial genome from various types of destructive external factors via in cellulo Dps–DNA co-crystallization. This protective response results in the emergence of bacterial resistance to antibiotics and other drugs. The protective properties of Dps have attracted a significant attention of researchers. However, Dps has another equally important functional role. Being a ferritin-like protein, Dps acts as an iron depot and protects bacterial cells from the oxidative damage initiated by the excess of iron. Here we investigated formation of iron oxide nanoparticles in the internal cavity of the Dps dodecamer. We used anomalous small-angle X-ray scattering as the main research technique, which allows to examine the structure of metal-containing biological macromolecules and to analyze the size distribution of metal nanoparticles formed in them. The contributions of protein and metal components to total scattering were distinguished by varying the energy of the incident X-ray radiation near the edge of the metal atom absorption band (the K-band for iron). We examined Dps specimens containing 50, 500, and 2000 iron atoms per protein dodecamer. Analysis of the particle size distribution showed that, depending on the iron content in the solution, the size of the nanoparticles formed inside the protein molecule was 2 to 4 nm and the growth of metal nanoparticles was limited by the size of the protein inner cavity. We also found some amount of iron ions in the Dps surface layer. This layer is very important for the protein to perform its protective functions, since the surface-located N-terminal domains determine the nature of interactions between Dps and DNA. In general, the results obtained in this work can be useful for the next step in studying the Dps phenomenon, as well as in creating biocompatible and solution-stabilized metal nanoparticles.
KEY WORDS: ferritin-like proteins, Dps, iron oxide, anomalous SAXS, size distributionDOI: 10.1134/S0006297922060037
Abbreviations: SAXS, small angle X-ray scattering.
INTRODUCTION
Approximately one-third of the well-characterized proteins and almost half of the enzymes contain from one to several metal ions [1, 2]. Among those, one of the most vital and abundant chemical elements is iron. This metal is commonly found in the hemes or iron-sulfur prosthetic groups in proteins [2, 3]. The biological significance of iron is determined by its ability to undergo reversible redox reactions. Divalent and trivalent iron is essential in vital processes in all eukaryotes and most prokaryotes. Iron participates in various metabolic processes, such as oxygen transport, DNA synthesis, and electron transport for energy production. It is a universal microelement that ensures normal functioning of all body systems at the cellular level.
On the other hand, the excess of iron promotes the damage of DNA, proteins, and lipids, leading to the disruption of cellular homeostasis. The existing evolutionary developed mechanisms of detoxification and iron removal from the cytosol include oxidation of excessive divalent iron to the trivalent iron in the Fenton reaction [4]:
In living organisms, trivalent iron is accumulated in the ferritin-like proteins. The superfamily of ferritin protein has evolved to provide iron sequestration in its soluble, non-toxic, and bioavailable form [5]. These proteins function as iron depots and store it in a non-reactive form, this limiting its involvement in cellular processes and protecting the cell from the oxidative damage initiated by this metal [6, 7]. Ferritin is a protein complex that plays a role of major intracellular iron depot in humans and animals. It is found virtually in all organs and tissues. Mammalian apoferritin (protein lacking iron ions) is a 24-mer protein with the molecular mass (MM) of ∼450 kDa; each of its polypeptide subunits has MM of ∼20 kDa. Ferritin is a globular protein with the inner cavity that can store approximately 4500 ions of hydrated trivalent iron oxide (Fe2O3 • H2O) together with a variable number of phosphate groups. The external diameter of the protein is 12-13 nm; the dimeter of its inner cavity is 7-8 nm [7, 8]. The protein component of the ferritin molecule has numerous pores through which iron is transported [9].
There are at least three types of ferritin-like prokaryotic proteins – bacterial ferritin, bacterioferritin, and dodecamer ferritin (mini-ferritin) – that are related to eukaryotic ferritins. Similar to the mammalian ferritins, bacterial ferritin and bacterioferritin consist of 24 subunits and have the central cavity that can accommodate ∼2500 iron atoms in bacterial ferritin [10] and ∼1800 iron atoms in bacterioferritin [10, 11]. The presence of heme (iron protoporphyrin IX) located between each two protein subunits and linked to methionine in each of these subunits is a characteristic feature of bacterioferritin that distinguishes it from other ferritin forms. As a result, the bacterioferritin molecule contains 12 heme groups, the role of which is yet to be elucidated [12-14].
Dps (DNA-binding protein from starved cells) has a special place among the dodecamer mini-ferritins. Its most important function is protection of genome from harmful factors such as starvation, high temperature, UV- and γ-radiation, toxins, chemical shock, and oxidative stress. Dps binds to DNA thus forming a stable Dps–DNA complex that protects DNA from damage [15-17]. One of the important consequences of this protective response is the emergence of bacterial resistance to antibiotics and other drugs. That is why formation of protective Dps–DNA complexes in the stress-induced bacterial cells has attracted a significant attention of many research groups in recent two decades. The most known works in this area are the studies by Minsky and colleagues, who were the first to prove this phenomenon experimentally [18-20].
In bacterial cells, Dps is present in minor amounts, and its synthesis is induced during the stationary phase of bacterial growth, during starvation, or under oxidative stress [21]. The Dps dodecamer lacks structural modules for the recognition of specific nucleotide sequences. It is assumed that its binding with the negatively charged sugar-phosphate DNA backbone occurs via electrostatic interactions with the lysine-enriched N-terminal domains of Dps monomers [15, 22-24]. However, the exact mechanism of Dps binding to DNA is still unknown [25].
Dps monomer is a polypeptide consisting of 167 amino acid residues (MM 18.7 kDa), that contains a highly conserved sequence of four α-helices [26]. The Dps dodecamer has MM 224.4 kDa and displays type 23 symmetry with the external diameter of 8-9 nm with the inner cavity diameter of 4-5 nm. As Dps is a ferritin-like protein, each monomer is capable of binding up to 40 iron atoms. Hence, Dps can accommodate approximately 500 Fe3+ ions in its inner cavity [27].
Therefore, Dps simultaneously performs two vitally important functions in bacterial cells: (i) protects bacterial genome from unfavorable external factors by forming a crystal complex with DNA; (ii) serves as an iron depot and protects cells from the oxidative damage initiated by the iron excess. These two functions are interrelated. In particular, Zhao et al. [27], who studied DNA damage in vitro, demonstrated that Dps prevents the cleavage of Escherichia coli DNA simultaneously exposed to FeSO4 and H2O2. That implies that by forming the protein–DNA complex and neutralizing hydrogen peroxide during interaction with iron, Dps preserves E. coli genome under stress conditions [28].
Despite the fact that E. coli Dps does not contain any canonical ferroxidase sites present in E. coli bacterioferritin or bacterial ferritin, Dps oxidizes Fe2+ at some sites in the protein macromolecule followed by its accumulation as Fe3+ in the protein central cavity [27]. It has been assumed that the ferroxidase sites of Dps are formed by the iron-binding amino acids, such as aspartate, glutamate, and histidine [28, 29]. Thus, the N-terminal domain of the expressed Deinococcus radiodurans Dps (DrDps1) contains a region (residues 30-55) that includes the metal-binding site (Asp36x2His39x10His50x4Glu55 motif) and is located at the external surface of the dodecamer. Disruption of this site affects the protein self-assembly, as well as reduces its DNA-binding capacity, i.e., decreases its protective capacity [30].
Unlike Dps, far from all bacterial dodecamer mini-ferritins can simultaneously protect an organism from the oxidative stress by forming a crystalline complex with DNA and serve as a source of iron in the case of its deficit [31]. That is one of the reasons why the unique features of Dps attract so much attention. Moreover, there is also another aspect that makes investigating this protein so important. Accumulation of a large number of metal ions in the inner cavity of Dps protein presumes an existence of magnetic properties; such protein molecules could be considered as natural biosensors of electromagnetic radiation. Dps is capable of transmitting the received signal to DNA, which gives a promise for the development of a new generation of logic elements. That is why investigating the formation of iron nanoparticles inside the stabilizing Dps protein shell has not only scientific, but also practical significance.
In order to use the small angle X-ray scattering (SAXS) for elucidating the structure of metal-containing specimens and size distribution of metal nanoparticles in the sample, the contribution of these components to the total scattering should be separated. Traditional SAXS techniques are used for studying the structure of original samples prior to the formation of metal particles (i.e., structure of the matrix) [32]. The standard approach based on the subtraction of the matrix scattering from the total scattering of the metal-containing sample is possible only in the cases, when the structure of the matrix does not change during formation of the metal particles in it. When structural changes in the original sample are presumed, the commonly used method is anomalous SAXS (ASAXS) [33-36]. In this method, experimental SAXS curves obtained for the metal-containing samples are recorded at different energies of the X-ray radiation: (i) close to the edge of the absorption band of the metal (when only matrix scattering is recorded) and (ii) away from the band edge (when total scattering is recorded), so that the difference between these two curves could be attributed only to the scattering of nanoparticles containing the metal atoms. Analysis of the contribution of each component allows to calculate the size distribution of the metal nanoparticle and to evaluate the structure of the metal-containing sample during its interaction with the metal.
In this study, we investigated accumulation of iron oxide in the inner cavity of the ferritin-like Dps protein using ASAXS.
MATERIALS AND METHODS
Preparation of iron-containing Dps samples. Dps was expressed and purified according as described earlier [37, 38]. The purified protein was concentrated with an Amicon® centrifugal concentrator (10 kDa cut-off; Merck Millipore, USA) to the concentration of 3 mg/ml followed by dialysis against a buffer containing 50 mM NaCl, 0.5 mM EDTA, 50 mM Tris-HCl (pH 8.0). The purity of the obtained protein sample was evaluated by electrophoresis in a 15% polyacrylamide gel; protein concentration was determined from absorbance at 280 nm (A280) using molar absorption coefficient from [39].
Freshly prepared solution of FeSO4 ⋅ 7H2O was added to the purified Dps (3 mg/ml) in a buffer containing 50 mM NaCl, 0.5 mM EDTA, and 50 mM Tris-HCl (pH 8.0) in the amounts corresponding to 50, 500, and 2000 iron atoms per dodecamer, and the mixture was incubated for 30 min at room temperature. The resulting samples are designated in the text as Dps-Fe50, Dps-Fe500, and Dps-Fe2000, respectively.
SAXS and analysis of the obtained data. Traditional SAXS. Dps structure was investigated by traditional SAXS with the Petra III synchrotron (DESY, Germany), beamline P12 [40]. The P12 beamline was equipped with an automatic sample changer and two-dimensional Pilatus 2M detector (DECTRIS, Switzerland). The scattering intensity, I(s), was measured in the range of momentum transfer values 0.08 < s < 3 nm–1, where s is scattering vector (4πsinθ/λ), 2θ is the scattering angle, and λ is the radiation wavelength (0.124 nm). For each sample, 50 frames were recorded to evaluate possible radiation damage. No radiation damage was observed in the process.
The scattering curves were primarily processed with the PRIMUS program [41]. Analysis of the obtained data and structural modeling were performed using the ATSAS software package [42].
The radius of gyration (Rg) of the scattering particles was determined from the initial part of the scattering curve in the region of the smallest s values using the equation (1):
which is valid in the region (sRg) < 1.3. The scattering intensity at the zero angle, I(0), was determined from the slope of the linear part of the Guinier plot (ln I(s) versus s2), which is proportional to molecular mass of the scattering object.
Molecular masses were calculated from the SAXS data by two different methods: (i) using Bayesian approach (MMBayesian) [43] and (ii) based on the excluded volume Vp (Porod volume) inaccessible to the solvent [44] using the empirical ratio between Vp and MM (1.65 for proteins) [45].
The GNOM program [46] was used to calculate the distance distribution function p(r), which was required for the reconstruction of the Dps protein shape in solution based on the SAXS data. The distance distribution function, p(r), was determined as an indirect Fourier transform of the scattering intensity in accordance with the equation (2):
where I(s) is the scattering intensity. The maximum particle size (Dmax) was calculated under conditions p(r) = 0 at r > Dmax.
The ab initio method for the reconstruction of the low-resolution shape of Dps was based on the simulated annealing algorithm and was implemented using the DAMMIN program [47], which allows the construction of structural models with the minimization of residual χ2 between the experimental scattering and scattering obtained for the models (3):
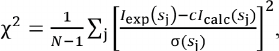 (3)
(3)where N is the number of experimental points, Iexp(sj) and σ(sj) are experimental intensities and errors, Icalc(sj) is the calculated intensity for the model, c is the scaling coefficient.
The structure of the iron-containing protein was calculated using the multiphase ab initio modeling, which allowed to obtain not only the low-resolution structure of the protein component in the complex, but also to determine the location of iron atoms in the protein matrix. The two-component (two-phase) model was constructed with the MONSA program [47]. This program takes into account the differences in the electron densities of the protein and metal components of the complex, as well as the ratio between their volumes. SAXS curves from the original protein and protein containing metal atoms were used to produce the two-phase model of the protein–metal complex. The theoretical scattering intensity for the constructed models was calculated using the CRYSOL program [48].
ASAXS experiments were also carried out with a synchrotron Petra III, beamline P12, and involved recording of X-ray scattering curves at different wavelengths (λ), i.e., at different energies of the incident beam (E). The measurements were conducted for the original Dps protein and Dps samples containing 50 (Dps-Fe50), 500 (Dps-Fe500), and 2000 (Dps-Fe2000) iron atoms per dodecamer. The scattering data were recorded at several different photon energies Ek with the E0 energy (10,000 keV, λ = 0.124 nm) being sufficiently far from the edge of the Fe absorption band, and, hence, selected for investigating the structure of the original Dps molecule (traditional SAXS technique). The obtained SAXS and ASAXS data were corrected for the background scattering and fluorescence, and were processed with the ATSAS software package [42] using the recently developed strategy for ASAXS data acquisition and processing [49].
The atomic scattering factor was determined from the following equation (4):
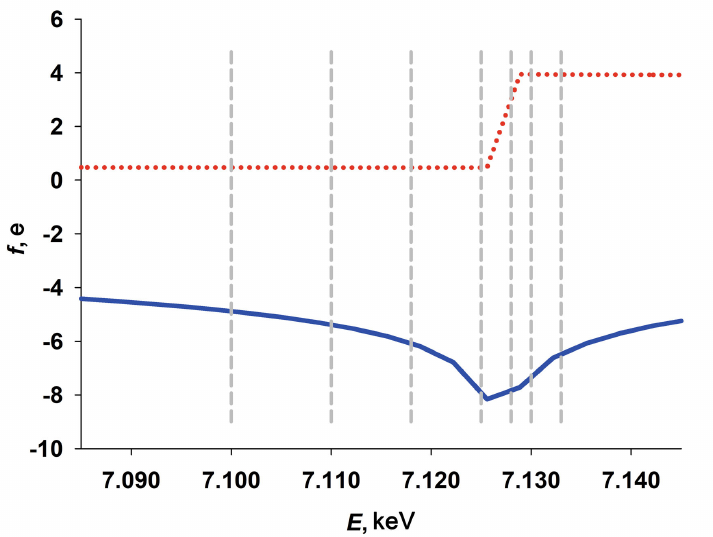
where dispersion correction factors f′(E) and f′′(E) become significant in the vicinity of the edge of the resonance atom absorption band. In our case, the measurements were conducted close to the K-band (absorption band of Fe atom), i.e., at the photon energy E = 7.125 keV (λ = 0.174 nm). The changes in the dispersion corrections factors f′(E) and f′′(E) with changes in the energy of photons used in this study, E, are shown in Fig. 1.
Fig. 1. Dependence of the real and imaginary components of ASAXS f′(E) (solid blue line) and f′′(E) (red dashed line) on the photon energy E close to the K-band (Fe atom absorption band) with E = 7.125 keV. The energy of radiation used for ASAXS measurements is shown by dashed vertical lines. The calculated values for the dispersion correction factors for the iron atoms were taken from the open access portal (http://skuld.bmsc.washington.edu/).
For each sample, the scattering curves I(s, Ek) were recorded with 7 different energies of the incident radiation E1-7: 7.100, 7.110, 7.118, 7.125, 7.128, 7.130, and 7.133 keV in the region at the edge of K-band (E = 7.125 keV). The anomalous correction factors for E = 7.125 keV were 8.13 and 0.48, respectively. The difference between the scattering curves produced at different energies ∆k(s) = I(s, E0) – I(s, Ek) is proportional to the scattering by the resonance atoms [33-36]. These difference curves were used for calculating the volume size distribution functions DV(R). The integral equation (5) was solved for DV(R) using the GNOM program [46] assuming a spherical shape of the formed nanoparticles.
In this equation, R is the sphere radius; Rmin and Rmax are the minimal and maximal sizes, respectively; i0(x) = {[sin(x) – x cos(x)]/x3}2 and m(R) = (4π/3)R3 are the sphere form factor and its volume, respectively. The scattering length density for anomalous atoms is defined as follows: ∆ρ = (N02 – Nk2)e/vat, where N0 and Nk are the number of electrons contributing to scattering far from the resonance and at E = Ek, respectively; e is the electron charge; and vat is atomic volume.
We also used an alternative approach to analyze the size distribution of Fe nanoparticles formed in the Dps protein using the MIXTURE program [41]. In this approach, the scattering intensity I(s) of the mixture of k different components with different sizes is represented as a linear combination (6):
where νk is the volume fraction of component k, Ik is the scattering intensity for this component, K is the number of components. The MIXTURE program models the scattering of mixtures containing K number of scattering objects with varying shape and size using theoretical scattering from the simple bodies (sphere, hollow sphere, ellipsoids, cylinders, etc.). Each object is characterized by its own volume fraction, average size, polydispersity distribution width, contrast, and, optionally, by the possibility of interparticle interactions. An experimental scattering pattern is approximated by the weighted combination of the calculated individual (partial) scattering curves from the components to minimize residual χ2 between the experimental and model data.
RESULTS AND DISCUSSION
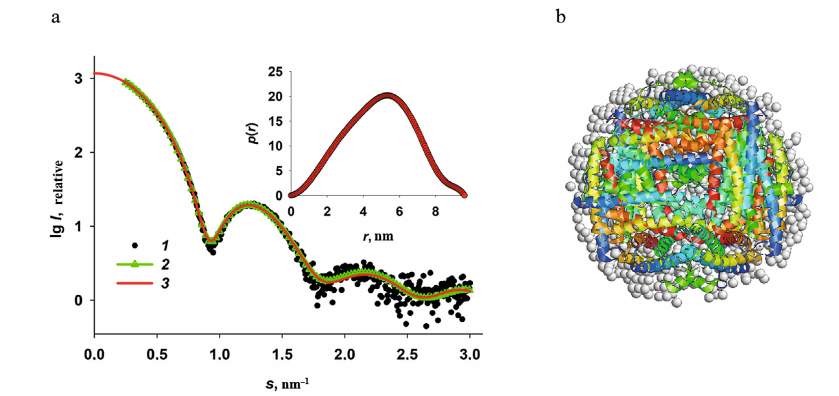
SAXS studies of the Dps structure. We used traditional SAXS technique to investigate the Dps structure in solution and to determine the SAXS invariants, such as radius of gyration (Rg), Porod volume (Vp), distance distribution function p(r), the maximum size (Dmax), and molecular mass (MM). Preliminary experiments have shown the absence of concentration dependence and aggregate formation in the solution at the Dps concentrations ranging from 1.0 to 3.0 mg/ml. Based on this observation, we selected the sample with the concentration of 3.0 mg/ml for further SAXS experiments and data processing, as it produced a sufficiently informative SAXS curve with a low noise in the range of angle vectors 0.25 < s < 3.0 nm–1 and clearly pronounced shape factor (Fig. 2, curve 1).
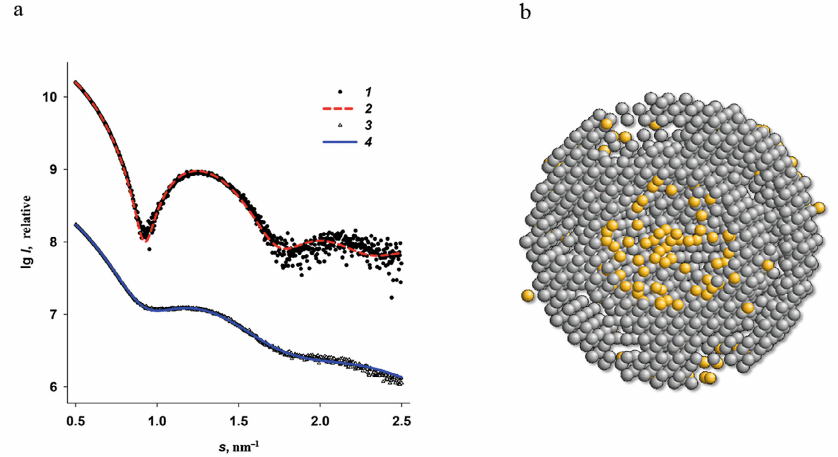
Fig. 2. Reconstruction of the low-resolution structure of Dps. a) Experimental SAXS curve (1); theoretical scattering curve calculated based on the bead model produced with the DAMMIN program (2); theoretical scattering curve calculated using the distance distribution function p(r). b) Superposition of the Dps crystal structure (colored helices) and the bead model obtained using the DAMMIN program (grey spheres).
The SAXS curve presented in Fig. 2 is typical for a solution of a monodisperse spherical protein. The distance distribution curve p(r) (Fig. 2, inset), which describes the shape of a scattering object [32] and was used in the DAMMIN program for reconstruction of the low-resolution structure from the SAXS data [47] also indicates a spherical shape of the protein. It can be concluded based on the p(r) function profile that in this case, the spherical object is hollow, since the distance distribution function is asymmetric and its maximum is shifted to the right. Moreover, considering that the amplitude of the p(r) function is proportional to the electron density of individual parts of a scattering object, it can be suggested that the protein has a less dense surface layer, because there is a weakly scattering “tail” in the p(r) profile in the size range of ∼8-9.6 nm. Based on the published data and our previous research, such scattering corresponds to the flexible N-terminal domains of the protein [24, 50, 51].
The low-resolution Dps structure (bead model) was reconstructed from the SAXS data using the DAMMIN program [47]. The shape reconstruction was based on the algorithm of annealing within the sphere with a diameter of the maximum size of the protein molecule, Dmax, which, in turn, was determined from the distance distribution function and was 9.6 nm. The results of the reconstruction are presented in Fig. 2b with grey beads. The residual χ2 for the experimental data was 1.9, which indicates good agreement of the experimental SAXS curve with the scattering curve produced by the obtained low-resolution shape (Fig. 2a, curve 2). The bead model of the Dps structure has an inner cavity and corresponds to the known crystal structure of the protein (PDB ID: 1DPS) (Fig. 2b), although the bead model is slightly bigger in size due to the scattering by the N-terminal domains (which are absent in the 1DPS atomic-resolution structure) due to their flexibility and impossibility of their crystallization [26].
The main structural characteristics of Dps determined directly from the SAXS curve without modeling (SAXS invariants) are presented in Table 1.
Table 1. Main structural characteristics of
Dps protein

The data in Table 1 together with the reconstructed low-resolution shape of the protein, indicate its native state, correspond to the characteristics of this protein known from the literature [26, 50, 51], and make it possible to further use the protein in investigating the processes of accumulation of iron atoms in the inner cavity of the Dps dodecamer.
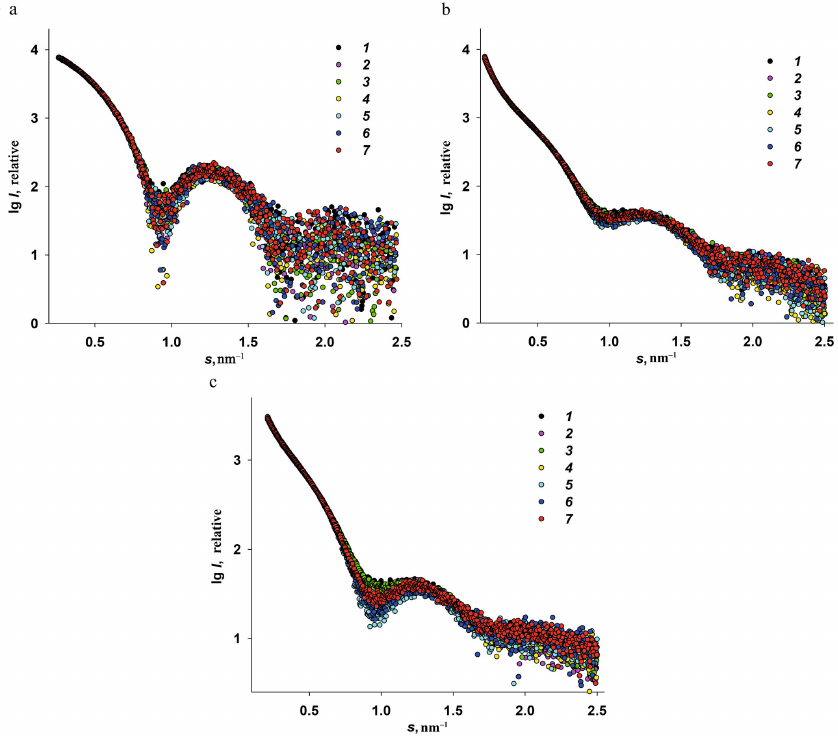
ASAXS studies of the iron nanoparticles formation in Dps. Experimental ASAXS curves recorded at different energies are presented in Fig. 3.
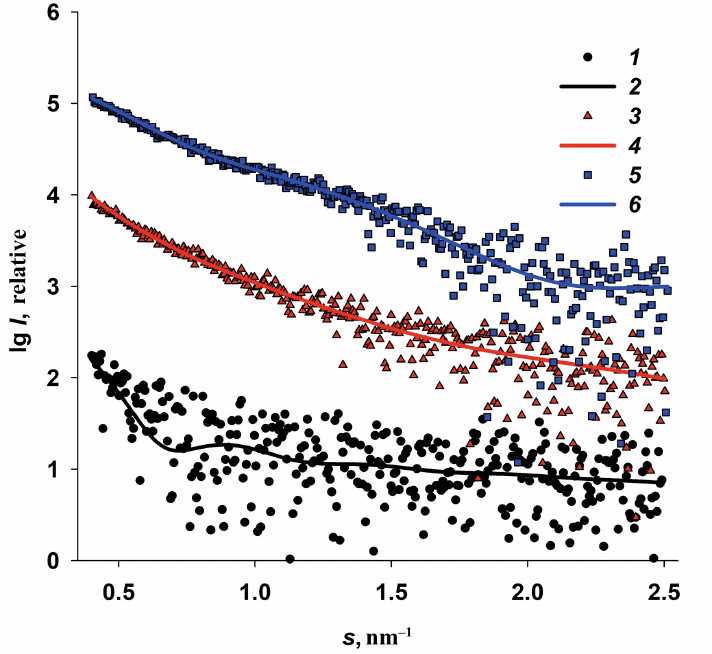
Fig. 3. Experimental ASAXS for three iron-containing Dps specimens recorded at three different photon energies: Dps-Fe50 (a); Dps-Fe500 (b); Dps-Fe2000 (c). Curve numbers (1-7) correspond to the radiation energies: 7.100, 7.110, 7.118, 7.125, 7.128, 7.130, and 7.133 keV, respectively.
Analysis of the scattering curves for the samples with different Fe3+ content and measured at different radiation energies E1-7 revealed the following features of the Dps interaction with iron ions.
1. The scattering curves recorded at the energy E0 in the regions far from the iron absorption band (10,000 keV, λ = 0.124 nm) and at the energies E1,2 (7.100 and 7.110 keV) were virtually identical for all Fe-containing samples. (Hence, we omitted SAXS curves recorded at E0 in Fig. 3 to avoid plot overloading.)
2. The low iron content (50 atoms per Dps molecule) did not affect the general structural characteristics of the protein at the energies relatively far from the iron absorption band (K-band). It is important to emphasize once more that the preliminary structural analysis of the original Dps protein at the energy E0 revealed that the protein was in the state suitable for its use as a matrix for the formation of iron nanoparticles.
3. In contrast to the specimens with the low iron content (Dps-Fe50), Dps-Fe500 and Dps-Fe2000 demonstrated a significant increase in the scattering intensity at very small angles, i.e., exhibit high polydisperse due to the ability of iron as a transition metal to form stable complexes with, for example, protein amino groups, thus bringing closer together protein chains of the neighboring macromolecules. The protein matrix, thus, changed significantly upon interaction with the iron-containing compounds, which strictly required the use of ASAXS in this case.
4. In the samples with the high iron content (Dps-Fe500 and Dps-Fe2000), the presence of large metal nanoparticles was expected due to the formation of metal-containing protein aggregates.
5. The scattering curves recorded at different photon energies E1-7 demonstrated a certain dependence on the energy of incident beam in the range of angle vectors 0.5 < s < 1.3 nm–1 with the minimum at s = 0.95 nm–1 (Fig. 3). Although this dependence was most pronounced for Dps-Fe2000, nevertheless, for all iron-containing samples it was possible to calculate the difference between the scattering curves obtained at different energies ∆k(s) = I(s, E1) – I(s, Ek), which was proportional to the scattering by the resonance Fe atoms and could be used for the analysis of size distribution of metal nanoparticles formed in the protein.
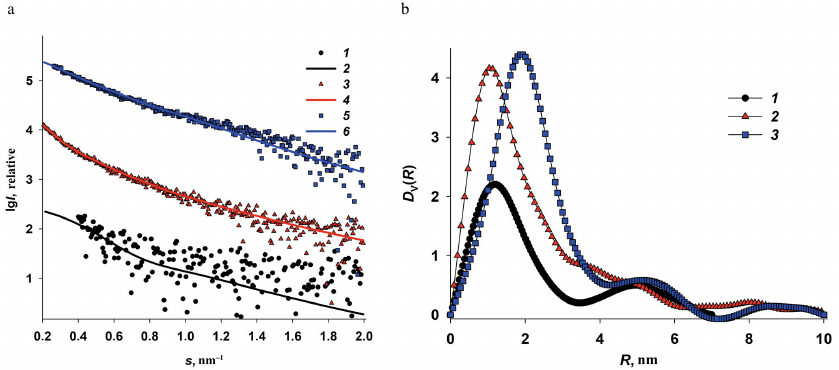
6. The maximum of the anomalous signal for all Fe-containing samples was observed at the photon energy E5 = 7.128 keV, which is close to the K-band of Fe absorption (Fig. 1). Accordingly, the scattering curve at this energy was used to evaluate the resonance signal ∆5(s) = I(s, E1) – I(s, E5) only from the iron nanoparticles formed in the Dps inner cavity. The criterion that the difference signal is indeed due to Fe particles is the absence of the signal from the protein matrix, i.e., from the Dps form factor, on the obtained anomalous scattering curves. This criterion was met for all Fe-containing samples (Fig. 4a).
Fig. 4. Determination of size distributions of iron oxide nanoparticles from the ASAXS data. a) Difference curves (anomalous signal) Δ 5(s) = I(s, E1) – I(s, E5) and the corresponding scattering curves calculated from nanoparticle size distributions for Dps-Fe50 (1 and 2); Dps-Fe500 (3 and 4), and Dps-Fe2000 (5 and 6 ). b) Volume size distributions DV(R) of the iron nanoparticles in Dps-Fe50 (1), Dps-Fe500 (2), and Dps-Fe2000 (3) calculated from the anomalous signal curves by the GNOM program.
The obtained curves of the anomalous signal were then used to calculate the volume size distribution functions DV(R) of iron oxide nanoparticles formed in the internal cavities of the Dps dodecamer. (Fig. 4b). It is important to note that the ASAXS curves were well pronounced for Dps-Fe500 and Dps-Fe2000, but not for Dps-Fe50 (weak signal, high noise). Hence, the size distribution function calculated from this curve should be considered only as an estimate.
Analysis of the volume size distribution functions (Fig. 4b) revealed the following regularities. Predominantly small nanoparticles ∼2-nm in size were formed at low iron ion concentrations. The increase in the iron content to 500 atoms per protein molecule resulted in the additional emergence of 4 to 5-nm particles (shoulder in the DV(R) function), as well as minor amounts of larger structures (up to 20 nm) associated with the iron oxide nanoparticles in protein aggregates. At the iron content of 2000 atoms per dodecamer, mostly 4 to 5-nm particles were formed, as well as large Fe-containing structures, while the 2-nm particles were absent. This implies that at the high iron concentration, the entire inner cavity of the protein is filled with metal nanoparticles. Their size is limited by the size of the Dps dodecamer inner cavity and determined by the iron concentration in the solution, which is important when using this protein matrix for the formation of solution-stabilized metal nanoparticles.
Similar results were obtained using an alternative approach for determining the fractional composition of iron nanoparticles in the protein dodecamer with the MIXTURE program, suggesting that the shape of the formed particles is spherical. For each specimen, three fractions with a broad size range (Dmin – Dmax) were specified. For each fraction, the average particle size and volume ratio of the fraction were calculated using the MIXTURE program (Table 2).
Table 2. Fraction compositional of iron
nanoparticles in Dps
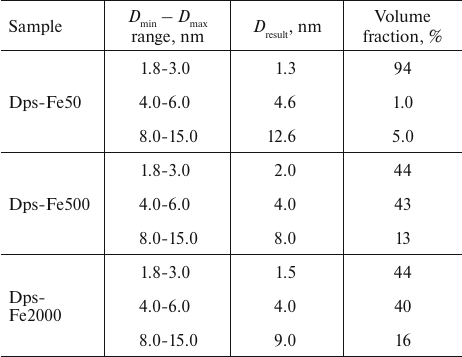
Similar to determining the volume size distribution DV(R) with the interactive GNOM program, the use of MIXTURE program revealed a predominant presence of the 2-4-nm nanoparticles in all samples together with a small amount of larger formations. Since both methods were used to determine the volume fractions of the nanoparticles, in quantitative terms the number of large particles was low even for the high iron concentration in the initial solution, but these large particles significantly contributed to SAXS due to their size (see Fig. 3, b and c).
The SAXS curves calculated from the fractional composition of each sample presented in Table 2 are in good agreement with the obtained anomalous signals (Fig. 5).
Fig. 5. Comparison of differential curves (anomalous signals) ∆5(s) = I(s, E1) – I(s, E5) and corresponding scattering curves based on the size distribution of iron-containing nanoparticles in Dps-Fe50 (1 and 2), Dps-Fe500 (3 and 4), and Dps-Fe2000 (5 and 6) and calculated with the MIXTURE program.
Multiphase ab initio modeling based on the SAXS data. According to the published data, the iron-binding sites are located not only in the inner cavity of the Dps protein, but also at the surface of the Dps dodecamer [28-30, 52]. Although the nanoparticles do not form on the dodecamer surface, each monomer contains up to four amino acid residues capable of metal binding in the region of the N-terminal domains, so the bound iron atoms contribute to the scattering of the entire macromolecule and to the size distribution, in particular, due to their high electron density. The ab initio multiphase modeling (MONSA program) is a SAXS technique that allows to use the difference between the electron density of different parts of the scattering object to localize some structural features associated with this difference [47]. The calculations are based on the differences between the electron density of the protein and metal components of the complex, as well as the ratio between the volumes of these components. In order to obtain a two-phase protein–metal model, we used the original SAXS curves of the iron-free and those of the iron-containing protein. We selected the Dps-Fe500 specimen for the modeling, which provided a reliable detection of the formed nanoparticles in the protein inner cavity, but at the same time, allowed localization of the significantly smaller metal-containing groups on the dodecamer surface on the background of strong scattering by the large particles. The results of modeling are presented in Fig. 6.
Fig. 6. Multiphase modeling of the Dps complex with iron using the MONSA program. a) Experimental SAXS curve for the Dps protein (1); scattering by the protein part of the multiphase model (2); experimental data for the Dps-Fe500 complex (3); scattering by the entire multiphase model (4). b) Multiphase model. Grey beads represent protein, yellow spheres are iron clusters. The cross-section of the model is shown for better visualization.
The multiphase modeling has allowed for the first time to visualize the location of iron atoms in the protein surface layer, and, at the same time, to confirm the presence of most metal ions in the central cavity of the Dps dodecamer. We observed a good agreement between the experimental data and calculated curves for the bead models with χ2 = 2.1. This generalized model provides the most comprehensive picture of formation of iron oxide nanoparticles in the ferritin-like protein that also protects bacterial genome.
CONCLUSIONS
Not all mini-ferritins protect bacterial genomes during oxidative stress, and not all nucleotide-associated proteins are capable of detoxification and accumulation of iron atoms. Hence, the multifunctionality of the ferritin-like protein Dps is unique, and its consequence is the stable resistance of bacteria to drugs and antibiotics. Undoubtfully, the main feature of Dps, which attracts a considerable interest of researchers, is its ability for the in cellulo biocrystallization with DNA. The persistent microbial cell formed via this mechanism are resistant to numerous adverse environmental factors; they can retain their viability for a long time and give rise to a new population with preserved pathogenic properties under favorable conditions [53]. That is why DNA “archiving” by in cellulo co-crystallization with Dps requires attention and detailed investigation. However, it is equally important to study the ability of Dps for detoxification and transformation of toxic Fe2+ into non-toxic Fe3+ ions followed by the accumulation of the trivalent iron in the protein inner cavity that is protected from the external exposure. These both functions of Dps are closely interrelated [30]. In our previous studies, we have investigated the formation of the Dps–DNA co-crystals, in particular, those formed with the involvement of divalent metal ions [37, 38, 50]. In this work, we studied the second feature of the functional properties of Dps, namely, the accumulation of iron ions in the inner cavity of this mini-ferritin. Using ASAXS technique, we were able to demonstrate that 2 to 4-nm nanoparticles were basically formed within the protein molecule, indicating that the growth of metal nanoparticles was limited by the spatial characteristics of the protein inner cavity. However, a certain number of iron ions was found in the Dps surface layer. This layer is very important for the protective functions of this protein, because the N-terminal domains located in this layer define the type of interactions of Dps with DNA in solution, since these flexible lysine-enriched N-terminal domains are responsible for the interaction with the negatively charged sugar-phosphate backbone of deoxyribonucleic acid [15, 22-24]. Therefore, the N-terminal domains must be accessible to bind DNA [50, 51, 53]. However, iron atoms could bind to the negatively charged amino acids (Asp and Glu) in the same N-terminal regions of Dps. Iron is a transition metal and can form stable complexes, for example, with the amino group nitrogen (N) in amino acids. Therefore, divalent iron ions could form bonds with the neighboring protein chains, pulling them towards each other and stabilizing the surface regions of the dodecamer. In this case, the N-terminal domains are pressed to the protein surface and the Dps–DNA crystal complexes are not formed [50, 54]. However, in the presence of chelating agent (e.g., EDTA), the bonds between the iron ions and amino acids on the dodecamer surface are disrupted, which can result in the restoration of the Dps interactions with DNA without affecting protein inner regions where the nanoparticles have formed. Hence, the charge of the iron ions in the composition of Dps (on the surface and in its core) is of great importance and requires detailed investigation.
SAXS structural studies do not allow to determine the valence of the iron atoms. By definition, the function of ferritin-like proteins, including Dps, is oxidation of Fe2+ to the trivalent state followed by accumulation of this non-toxic form in the protein central cavity. However, it was demonstrated that the mechanism of removal of the divalent iron from the solution and its oxidation at the respective protein sites is more complicated. In particular, the use of the Mössbauer spectroscopy for determination of the iron ion charge in Dps demonstrated that this charge was non-uniform. The protein contained iron in a form of magnetite FeO · Fe2O3, i.e., a mixture of di- and trivalent iron ions [55]. The authors concluded that this composition of the Dps inorganic core supports its dodecamer structure.
In general, formation of nanoparticles that contain metal atoms, and not only iron, but, for example, cobalt and possibly others, in the Dps inner cavity is of certain practical interest. Hence, the studies carried out in this work can be useful not only in terms of studying the unique properties of Dps, but can also contribute to the development of solution-stabilized biocompatible nanocapsules with magnetic properties. The plasticity and structural stability of the Dps protein matrix provides this possibility [56].
Acknowledgments. The authors are grateful to A. Gruzinov, European Molecular Biology Laboratory (EMBL), for conducting SAXS experiments and valuable discussion.
Funding. This work was supported by the Russian Science Foundation (project no. 18-74-10071).
Ethics declarations. The authors declare no conflicts of interest in financial or any other sphere. This article does not contain description of studies with human participants or animals performed by any of the authors.
Open access. This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution, and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
REFERENCES
1.Waldron, K. J., Rutherford, J. C., Ford, D., and
Robinson, N. J. (2009) Metalloproteins and metal sensing,
Nature, 460, 823-830.
2.Andreini, C., Bertini, I., Cavallaro, G., Holliday,
G. L., and Thornton, J. M. (2008) Metal ions in biological catalysis:
from enzyme databases to general principles, J. Biol. Inorg.
Chem., 13, 1205-1218.
3.Beinert, H., Holm, R. H., and Munck, E. (1997)
Iron-sulfur clusters: nature’s modular, multipurpose structures,
Science, 277, 653-659.
4.Fenton, H. J. H. (1894) Oxidation of tartaric acid
in presence of iron, J. Chem. Soc. Trans., 65,
899-910.
5.Briat, J.-F. (1992) Iron assimilation and storage
in prokaryotes, J. Gen. Microbiol., 138, 2475-2483.
6.Andrews, S. C. (1998) Iron storage in bacteria,
Adv. Microbial. Physiol., 40, 281-351.
7.Harrison, P. M., and Arosio, P. (1996) The
ferritins: Molecular properties, iron storage function and cellular
regulation, Biochim. Biophys. Acta, 1275, 161-203.
8.Theil, E. C. (1987) Ferritin, structure, gene
regulation, and cellular function in animals, plants, and
microorganisms, Annu. Rev. Biochem., 56, 289-315.
9.Levi, S., Salfeld, J., Franceschinelli, F., Cozzi,
A., Dorner, M. H., et al. (1989) Expression and structural and
functional properties of human ferritin L-chain from Escherichia
coli, Biochemistry, 28, 5179-5184.
10.Hudson, A. J., Andrews, S. C., Hawkins, C.,
Williams, J. M., Izuhara, M., et al. (1993) Overproduction,
purification and characterization of the Escherichia coli
ferritin, Eur. J. Biochem., 218, 985-995.
11.Andrews, S. C., Smith, J. M. A., Guest, J. R.,
and Harrison, P. M. (1990) Genetic and structural characterization of
the bacterioferritin of Escherichia coli, Biochem.
Soc. Trans., 18, 658-659.
12.Cheesman, M. R., Le Brun, N. E., Kadir, F. H. A.,
Thomson, A. J., Moore, G. R., et al. (1993) Haem and non-haem iron
sites in Escherichia coli bacterioferritin: Spectroscopic
and model building studies, Biochem. J., 292, 47-56.
13.Chasteen, N. D. (1998) Ferritin. Uptake, storage,
and release of iron, Met. Ions Biol. Syst., 35,
479-514.
14.Romão, C. V., Louro, R., Timkovich, R.,
Lübben, M., Liu, M.-Y., et al. (2000) Iron-coproporphyrin III is a
natural cofactor in bacterioferritin from the anaerobic bacterium
Desulfovibrio desulfuricans, FEBS Lett.,
480, 213-216.
15.Almiron, M., Link, A. J., Furlong, D., and
Kolter, R. (1992) A novel DNA-binding protein with regulatory and
protective roles in starved Escherichia coli, Genes
Dev., 6, 2646-2654.
16.Nair, S., and Finkel, S. E. (2004) Dps protects
cells against multiple stresses during stationary phase, J.
Bacteriol., 186, 4192-4198.
17.Frenkiel-Krispin, D., and Minsky, A. (2006)
Nucleoid organization and the maintenance of DNA integrity in E.
coli, B. subtilis and D. radiodurans, J. Struct.
Biol., 156, 311-319.
18.Frenkiel-Krispin, D., Ben-Avraham, I., Englander,
J., Shimoni, E., Wolf, S. G., et al. (2004) Nucleoid restructuring in
stationary-state bacteria, Mol. Microbiol., 51,
395-405.
19.Reich, Z., Wachtel, E. J., and Minsky, A. (1994)
Liquid-crystalline mesophases of plasmid DNA in bacteria,
Science, 264, 1460-1463.
20.Wolf, S. G., Frenkiel, D., Arad, T., Finkel, S.
E., Kolter, R., et al. (1999) DNA protection by biocrystallization,
Nature, 400, 83-85.
21.Stephani, K., Weichart, D., and Hengge, R. (2003)
Dynamic control of Dps protein levels by ClpXP and ClpAP proteases in
Escherichia coli, Mol. Microbiol., 49,
1605-1614.
22.Azam, T. A., and Ishihama, A. (1999) Twelve
species of the nucleoid-associated protein from Escherichia
coli, J. Biol. Chem., 274, 33105-33113.
23.Azam, T. A., Iwata, A., Nishimura, A., Ueda, S.,
and Ishihama, A. (1999) Growth phase-dependent variation in protein
composition of the Escherichia coli nucleoid, J.
Bacteriol., 181, 6361-6370.
24.Ceci, P., Cellai, S., Falvo, E., Rivetti, C.,
Rossi, G. L., et al. (2004) DNA condensation and self-aggregation of
Escherichia coli Dps are coupled phenomena related to the
properties of the N-terminus, Nucleic Acids Res., 32,
5935-5944.
25.Gupta, S., and Chatterji, D. (2003) Bimodal
protection of DNA by Mycobacterium smegmatis DNA-binding
protein from stationary phase cells, J. Biol. Chem., 278,
5235-5241.
26.Grant, R. A., Filman, D. J., Finkel, S. E.,
Kolter, R., and Hogle, J. M. (1998) The crystal structure of Dps, a
ferritin homolog that binds and protects DNA, Nat. Struct.
Biol., 5, 294-303.
27.Zhao, G., Ceci, P., Ilari, A., Giangiacomo, L.,
Laue, T. M., et al. (2002) Iron and hydrogen peroxide detoxification
properties of DNA-binding protein from starved cells. A ferritin-like
DNA-binding protein of Escherichia coli, J. Biol.
Chem., 277, 27689-27696.
28.Ilari, A., Ceci, P., Ferrari, D., Rossi, G. L.,
and Chiancone, E. (2002) Iron incorporation into Escherichia
coli Dps gives rise to a ferritin-like microcrystalline core,
J. Biol. Chem., 277, 37619-37623.
29.Ilari, A., Stefanini, S., Chiancone, E., and
Tsernoglou, D. (2000) The dodecameric ferritin from Listeria
innocua contains a novel intersubunit iron-binding site, Nat.
Struct. Biol., 7, 38-43.
30.Nguyen, K. H., and Grove, A. (2012) Metal binding
at the Deinococcus radiodurans Dps-1N-terminal metal site
controls dodecameric assembly and DNA binding, Biochemistry,
51, 6679-6689.
31.Smith, J. L. (2004) The physiological role of
ferritin-like compounds in bacteria, Crit. Rev. Microbiol.,
30, 173-185.
32.Feigin, L. A., and Svergun, D. I. (1987)
Structure Analysis by Small-Angle X-Ray and Neutron Scattering, Plenum
Press, New York.
33.Stuhrmann, H. B. (1980) Anomalous dispersion of
small-angle scattering of horse-spleen ferritin at the Iron K
absorption edge, Acta Crystallogr. A, 36, 996-1001.
34.Stuhrmann, H. B., and Notbohm, H. (1981)
Configuration of the four iron atoms in dissolved human hemoglobin as
studied by anomalous dispersion, Proc. Natl. Acad. Sci. USA,
78, 6216-6220.
35.Stuhrmann, H. B. (1985) Resonant scattering in
macromolecular structure research, Adv. Polym. Sci., 67,
123-163.
36.Stuhrmann, H. B., Goerigk, G., and Munk, B.
(1994) Anomalous X-ray Scattering, Elsevier, Amsterdam.
37.Dadinova, L. A., Chesnokov, Y. M., Kamyshinsky,
R. A., Orlov, I. A., Petoukhov, M. V., et al. (2019) Protective
Dps–DNA co-crystallization in stressed cells: An in
vitro structural study by small-angle X-ray scattering and
cryo-electron tomography, FEBS Lett., 593, 1360-1371.
38.Kamyshinsky, R., Chesnokov, Y., Dadinova, L.,
Mozhaev, A., Orlov, I., et al. (2020) Polymorphic protective
Dps–DNA co-crystals by cryo electron tomography and small angle
X-ray scattering, Biomolecules, 10, 39.
39.Grimsley, G. R., and Pace, C. N. (2004)
Spectrophotometric determination of protein concentration, Curr.
Protoc Protein Sci., 3, doi:
10.1002/0471140864.ps0301s33.
40.Blanchet, C. E., Spilotros, A., Schwemmer, F.,
Graewert, M. A., Kikhney, A., et al. (2015) Versatile sample
environments and automation for biological solution X-ray scattering
experiments at the P12 beamline (PETRA III, DESY), J. Appl.
Crystallogr., 48, 431-443.
41.Konarev, P. V., Volkov, V. V., Sokolova, A. V.,
Koch, M. H. J., and Svergun, D. I. (2003) PRIMUS: A Windows PC-based
system for small-angle scattering data analysis, J. Appl. Cryst.,
36, 1277-1282.
42.Manalastas-Cantos, K., Konarev, P. V., Hajizadeh,
N. R., Kikhney, A. G., Petoukhov, M. V., et al. (2021) ATSAS
3.0: Expanded functionality and new tools for small-angle scattering
data analysis, J. Appl. Cryst., 54, 343-355.
43.Hajizadeh, N. R., Franke, D., Jeffries, C. M.,
and Svergun, D. I. (2018) Consensus Bayesian assessment of protein
molecular mass from solution X-ray scattering data, Sci. Rep.,
8, 7204.
44.Porod, G. (1982) Small Angle X-Ray
Scattering, Academic Press, London.
45.Franke, D., Petoukhov, M. V., Konarev, P. V.,
Panjkovich, A., Tuukkanen, A., et al. (2017) ATSAS 2.8: A comprehensive
data analysis suite for small-angle scattering from macromolecular
solutions, J. Appl. Cryst., 50, 1212-1225.
46.Svergun, D. I. (1992) Determination of the
regularization parameter in indirect-transform methods using perceptual
criteria, J. Appl. Cryst., 25, 495-503.
47.Svergun, D. I. (1999) Restoring low resolution
structure of biological macromolecules from solution scattering using
simulated annealing, Biophys. J., 76, 2879-2886.
48.Svergun, D., Barberato, C., and Koch, M. H. J.
(1995) CRYSOL – a program to evaluate X-ray solution scattering
of biological macromolecules from atomic coordinates, J. Appl.
Cryst., 28, 768-773.
49.Gruzinov, A. Y., Schroer, M. A.,
Manalastas-Cantos, K., Kikhney, A. G., Hajizadeh, N. R., et al. (2021)
Anomalous SAXS at P12 beamline EMBL Hamburg: Instrumentation and
applications, J. Synch. Rad., 28, 812-823.
50.Soshinskaya, E. Yu., Dadinova, L. A., Mozhaev, A.
A., and Shtykova, E. V. (2020) Effect of buffer composition on
conformational mobility of N-terminal fragments of Dps and type of
interaction with DNA. Investigation with small-angle X-ray scattering
technique, Kristallografiya, 65, 886-895.
51.Dubrovin, E. V., Dadinova, L. A., Petoukhov, M.
V., Soshinskaya, E. Yu., Mozhaev, A. A., et al. (2021) Spatial
organization of Dps and DNA–Dps complexes, J. Mol. Biol.,
433, 166930.
52.Minato, T., Teramoto, T., Kakuta, Y., Ogo, S.,
and Yoon, K. S. (2020) Biochemical and structural characterization of a
thermostable Dps protein with His-type ferroxidase centers and outer
metal-binding sites, FEBS Open Bio, 10, 1219-1229.
53.Harms, A., Maisonneuve, E., and Gerdes, K. (2016)
Mechanisms of bacterial persistence during stress and antibiotic
exposure, Science, 354, aaf4268.
54.Dadinova, L., Kamyshinsky, R., Chesnokov, Yu.,
Mozhaev, A., Matveev, V., et al. (2021) Structural rearrangement of
Dps–DNA complex caused by divalent Mg and Fe cations, Int. J.
Mol. Sci., 22, 6056.
55.Antipov, S., Turishchev, S., Purtov, Yu.,
Shvyreva, U., Sinelnikov, A., et al. (2017) The oligomeric form of the
Escherichia coli Dps protein depends on the availability of iron
ions, Molecules, 22, 1904.
56.Zeth, K., Sancho-Vaello, E., and Okuda, M. (2019)
Metal positions and translocation pathways of the dodecameric
ferritin-like protein Dps, Inorg. Chem., 58,
11351-11363.