REVIEW: Astrocyte Activation Markers
Yana V. Gorina1,2,a*, Alla B. Salmina1,2,3, Alexander. I. Erofeev1, Evgeniy I. Gerasimov1, Anastasia V. Bolshakova1, Pavel M. Balaban1,4, Ilya B. Bezprozvanny1,5, and Olga L. Vlasova1
1Laboratory of Molecular Neurodegeneration, Peter the Great St. Petersburg Polytechnic University, 194091 St. Petersburg, Russia2Research Institute of Molecular Medicine and Pathobiochemistry, Voino-Yasenetsky Krasnoyarsk State Medical University, 660022 Krasnoyarsk, Russia
3Laboratory of Neurobiology and Tissue Engineering, Brain Institute, Research Center of Neurology, 105064 Moscow, Russia
4Laboratory of Cellular Neurobiology of Learning, Institute of Higher Nervous Activity, 117485 Moscow, Russia
5Department of Physiology, University of Texas Southwestern Medical Center, 75390 Dallas, USA
* To whom correspondence should be addressed.
Received May 18, 2022; Revised June 27, 2022; Accepted June 28, 2022
Astrocytes are the most common type of glial cells that provide homeostasis and protection of the central nervous system. Important specific characteristic of astrocytes is manifestation of morphological heterogeneity, which is directly dependent on localization in a particular area of the brain. Astrocytes can integrate into neural networks and keep neurons active in various areas of the brain. Moreover, astrocytes express a variety of receptors, channels, and membrane transporters, which underlie their peculiar metabolic activity, and, hence, determine plasticity of the central nervous system during development and aging. Such complex structural and functional organization of astrocytes requires the use of modern methods for their identification and analysis. Considering the important fact that determining the most appropriate marker for polymorphic and multiple subgroups of astrocytes is of decisive importance for studying their multifunctionality, this review presents markers, modern imaging techniques, and identification of astrocytes, which comprise a valuable resource for studying structural and functional properties of astrocytes, as well as facilitate better understanding of the extent to which astrocytes contribute to neuronal activity.
KEY WORDS: astrocytes, markers, imaging methods, miRNA, optogeneticsDOI: 10.1134/S0006297922090012
Abbreviations: AQP4, aquaporin 4; CaM, calmodulin; ChR2, channelrhodopsin-2; Cx, connexin; EGFP, enhanced green fluorescent protein; GFP, green fluorescent protein; GFAP, glial fibrillar acidic protein; GS, glutamine synthetase; NDRG2, N-myc downstream-regulated gene 2.
INTRODUCTION
Astrocytes have long been considered as auxiliary cells solely ensuring trophic, metabolic, and structural support to neurons [1]. However, over the last three decades extensive studies demonstrated that they play a pivotal role in physiological and pathological functions of the brain. In particular, astrocytes regulate ion homeostasis, cerebral blood flow, contribute to neovascularization, protect from excitotoxic neuronal injury and death, promote synapse formation, and participate in cerebral energy metabolism and development of the blood-brain barrier (BBB) [2].
Moreover, along with the pre- and post-synaptic membranes astrocytes form the so-called tripartite synapse thereby ensuring efficient synaptic transmission [3]. Upon that, astrocytes secrete gliotransmitters, cytokines, and metabolites participating in the neuron-astroglia coupling and gliovascular control [4].
In addition, it is also worth noting that astrocytes are able to modulate microglia phenotype and phagocytosis via crosstalk between astrocytes and microglia implying release of diverse signaling molecules by both cell types allowing to establish an autocrine feedback loop for targeted reciprocal modulation upon damage of the central nervous system (CNS) [5].
Overall, astrocytes represent a crucial element in the cerebral development and functioning [6]. Astrocyte activity is based on the dynamic intra- and inter-cellular interactions regulating synaptic plasticity, interaction in neuronal circuits, as well as contributing to memory consolidation [7]. Being a pivotal component of astrocyte activity, fluctuations of the intracellular free Ca2+ could occur spontaneously or due to neurotransmitter-mediated activation of the NMDARs (N-methyl-D-aspartate receptors) and TRP (Transient receptor potential) channels as well as GPCRs (G-protein-coupled receptors) [8]. Such astrocyte signaling mediated by elevated cytosolic Ca2+ concentration ensures gliotransmitter release, neurovascular interaction, as well as presynaptic structural remodeling [9]. Recent evidence [10] demonstrates that the astroglial type 1 cannabinoid receptors (CB1) determine availability of the hippocampal synaptic D-serine (synaptic NMDAR agonist). Hence, it demonstrates that the astroglial CB1 receptors act as a key component underlying physiological object recognition memory consolidation in hippocampus. For instance, the CB1 gene ablation results in the impaired D-serine binding to hippocampal synaptic NMDAR in the GFAP-CB1-knockout mice, which is associated with the impaired object recognition memory as well as decreased long-term potentiation at hippocampal synapses. It suggests a novel mechanism for astroglial regulation of synaptic plasticity via the D-serine-dependent control of NMDARs [10].
It should be also noted that heterogeneous gene expression, morphology, and functional interactions with local neuronal circuits is typical for astrocytes [11]. Neuron–astrocyte interaction is necessary for enabling diverse events such as synapse formation and functioning, neurotransmitter release and reuptake, production of neurophysiological cues, as well as control of neuronal survival. It must be mentioned in this regard that the functional neuron-astrocyte crosstalk is largely sustained both in adult and aging brain of the healthy animals, but becomes profoundly altered in the process of development of neurodegenerative diseases [12].
Obviously, visualization and identification of astrocytes, especially in the in situ specimens and in the brain in vivo is a challenging task. In this case, the problems are associated with the remarkable morphological heterogeneity and lack of the universal astrocyte marker able to reveal all astroglial lineage cells. Existing methods include classical histological staining and immunocytochemistry (performed on fixed tissues), genetically-controlled expression of the astrocyte-specific fluorescent markers, incubation with fluorescent probes, as well as intra-glial fluorescent dye injection.
Here, we discuss various specific molecular markers as well as current visualization and identification approaches for astrocytes characterized by polymorphism and multifunctionality, which might provide an opportunity to extend our understanding of the astrocyte contribution to neuronal activity, modulated synaptic transmission and plasticity, as well as bidirectional neuron-astrocyte crosstalk.
DIVERSITY OF ASTROCYTE MARKERS
Until recently, no universal marker able to stain and identify astrocytes in the CNS has been proposed, which is due to inherent morphological heterogeneity of astrocyte, which, in turn, coupled with the remarkably diverse expression of various molecules [13] (table).
Astrocyte markers
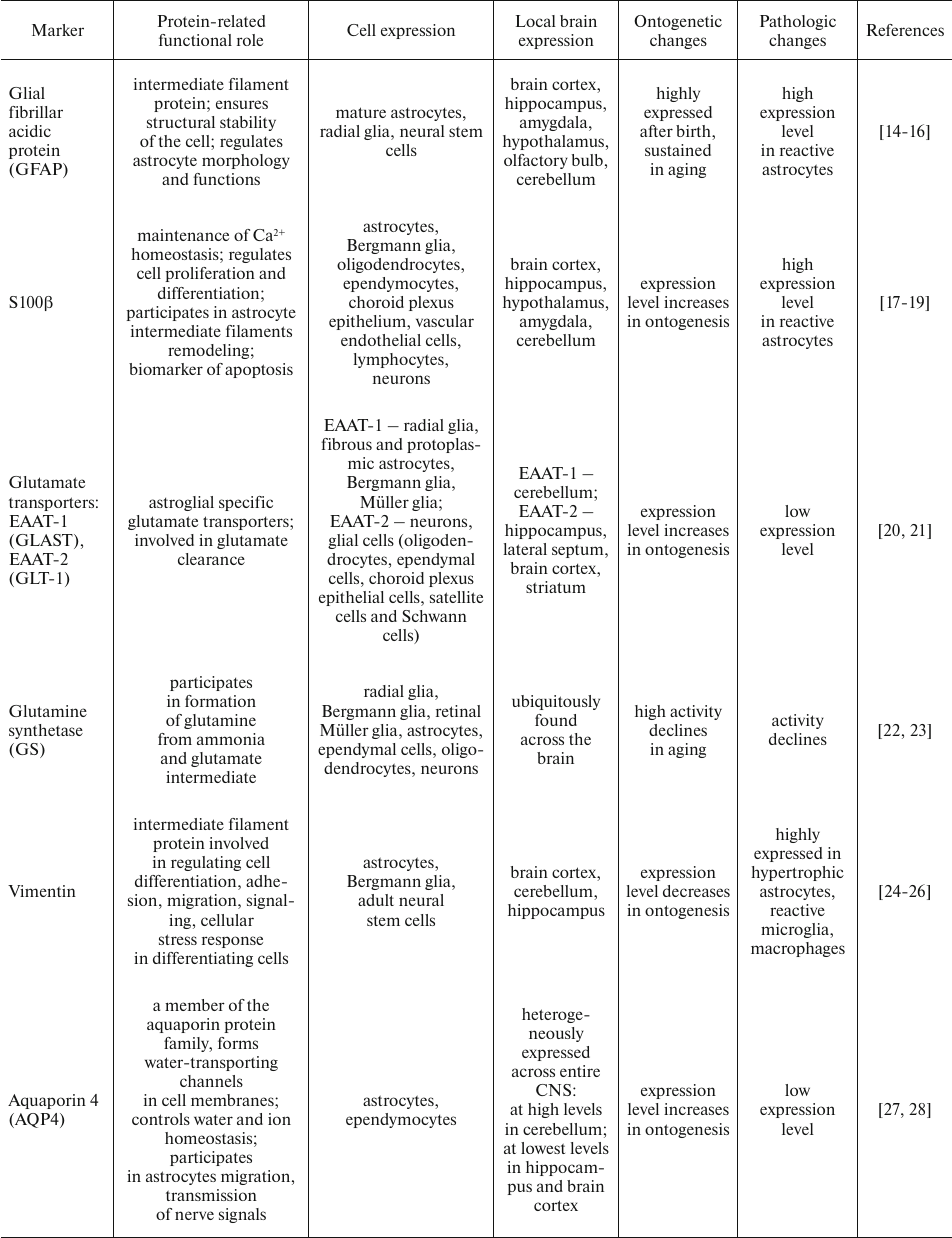
Glial fibrillar acidic protein (GFAP). Glial fibrillar acidic protein (GFAP) is one of the widely recognized astrocyte markers, which is the major intermediate filament protein found in mature astrocytes and crucial cytoskeletal constituent in the developing astrocytes [14]. It is worth emphasizing that GFAP as a common astrocyte-specific marker is mainly expressed in the white matter rather than in the grey matter astrocytes [44]. Moreover, GFAP is unable to fully delineate all astrocyte processes, 85% of which were negative for this marker [45].
Remarkably, under pathophysiological settings the upregulated GFAP expression serves as a sensitive and reliable reactive astrocyte marker, whereas in the intact tissue numerous astrocytes expressed it below the threshold of immunohistochemical detection, which, therefore, indicates that lack of the detected GFAP-positive signal does not allow to conclude that no astrocytes are present [16].
Moreover, it is also interesting that astrocytes in various cerebral areas may differ remarkably in the level of GFAP expression so that in the hippocampus, but not in the thalamus, it could be upregulated [14, 15].
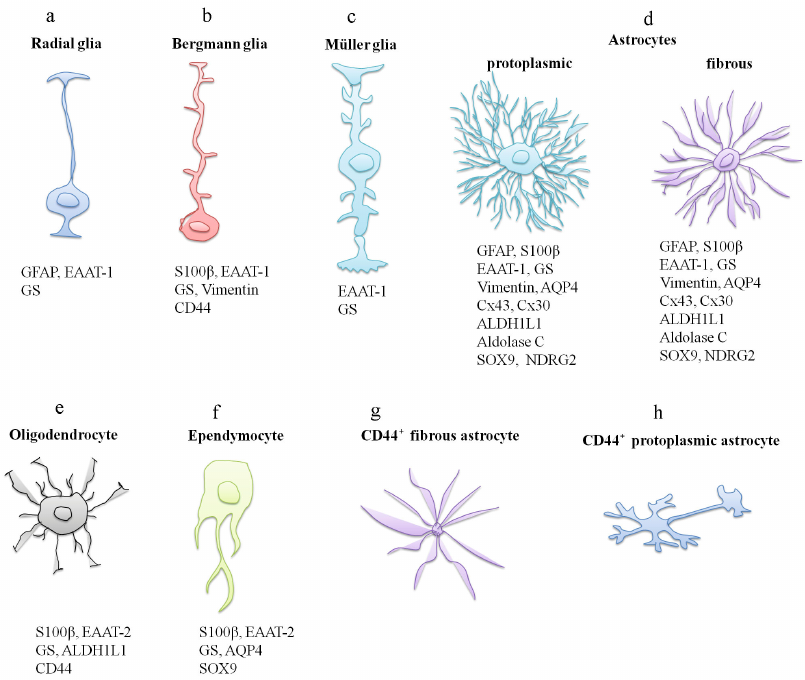
It was found that in the healthy human brain GFAP was mainly expressed in mature astrocytes [14]. However, experimental studies revealed that GFAP expression was observed in the radial glia of the developing human brain as well as adult neural stem cells [15, 46], thereby suggesting that GFAP is also expressed in immature undifferentiated cells in the CNS (figure).
Glial cells types and relevant visualization markers. a) Radial glia – GFAP expression [77], EAAT-1 [78], GS [77]; b) Bergmann glia – expression of S100β [79], EAAT-1 [80], GS [81], Vimentin [82], CD44 [41]; c) Müller glia – expression of EAAT-1 [83], GS [84]; d) astrocytes – expression of GFAP, S100β, EAAT-1, vimentin, AQP4, ALDH1L1 [85], GS [86], Cx43 [87], Cx30 [88], aldolase C [89], SOX9 [37], NDRG2 [90]; e) oligodendrocytes – expression of S100β [56], EAAT-2 [91], GS [92], ALDH1L1 [93], CD44 [94]; f) ependymocytes – expression of S100β [95], EAAT-2 [96], GS [77], AQP4 [97], SOX9 [98]; g) CD44+ fibrous astrocyte [75]; h) CD44+ protoplasmic astrocyte [75]
Furthermore, it was additionally demonstrated in the study aimed at investigating mouse cortical somatosensory cells and hippocampal CA1 region using the single-cell RNA sequencing (scRNA-Seq) [47] that they contained 47 molecularly distinct subclasses including all known major cell types in the cortex. Among them, astrocytes formed only two subclasses, one of which contained GFAP-positive astrocytes, whereas another – astrocytes expressing milk fat globule epidermal growth factor 8 (Mfge8), suggesting again limited GFAP expression in the substantial portion of astroglial cells. Moreover, it is important to take into account that GFAP is expressed in the radial glial cells (indeed being stem cells of the CNS), therefore, showing that GFAP detection in neurogenesis brain regions does not correspond to the position of mature astrocytes, which necessitates application of alternative markers.
S100β. Glycoprotein S100β is a Ca2+-binding protein regulating cell proliferation and differentiation [48]. It was shown that astrocytes isolated from the S100β-knockout mice exhibited markedly impaired Ca2+ signaling that indicated crucial role played by S100β in maintaining the Ca2+ homeostasis in astrocytes [49].
Noteworthy is the fact that S100β could participate in the astrocyte intermediate filament remodeling by preventing excessive GFAP polymerization in the presence of micromolar Ca2+ concentration, which, in turn, corroborates the idea that the astrocyte intermediate filaments represent dynamic cytoskeletal structures as well as existing cytosolic cues involved in regulating their assembly [50].
It is known that astrocytes produce and express S100β, which provides the expression-level-dependent neuroprotective or neurotoxic effect, stimulates astroglial cell proliferation by promoting its reactivation at higher level, as well as regulates microglia activation at inflammatory sites [51]. Moreover, experimental data suggest that S100β acts as a glial modulator of neuronal synaptic plasticity and long-term potentiation [52].
In pathological states, S100β expression becomes profoundly altered so that, e.g., its increased serum and cerebrospinal fluid level could be of some diagnostic importance in context of astroglial lineage cell damage [53].
S100β is used as an astrocyte marker due to its high expression level especially in perivascular astroglia. For instance, dual immunofluorescent staining for S100β and GFAP showed that as few as around 80% of the S100β-positive cells were simultaneously positive for GFAP-specific signal, whereas all GFAP-positive cells also co-expressed S100β in the mouse hippocampus. Upon that, the level of expression of S100β in astrocyte was markedly higher than of GFAP. Moreover, the GFAP-positive cells expressing S100β virtually did not differ in size and shape, as well as in S100β-specific immunoreactivity from the GFAP-negative cells [54] (figure).
Furthermore, comparative analysis of the distribution and expression of the three astrocyte-coupled markers such as NDRG2, GFAP, and S100β in the cortex, hippocampus, and thalamus revealed that in comparison with the GFAP-immunopositive astrocytes, NDRG2- and S100β-expressing astrocytes were distributed more evenly across the entire brain. High level of expressed NDRG2 and S100β was observed in the cortex and thalamus, whereas for GFAP it was found in the hippocampus [55].
In addition to this, S100β is expressed in astrocytes both in the white matter and grey matter [56], whereas GFAP was solely observed in the white matter [57]. However, the cell type-specificity of S100β expression was prominently inferior to that of GFAP. In particular, apart from astrocytes, S100β expression has been also observed in oligodendrocytes [17], ependymocytes [18], vascular endothelium, lymphocytes [19] as well as neurons [58] (figure).
S100a10 is a member of S100 protein family expressed in multiple organs (heart, kidneys, liver, lungs, spleen, gastrointestinal tract), including brain, where it plays an important role in transmembrane trafficking, vesicle secretion, and endocytosis [59]. Unlike the expression of S100β, the S100a10 expression was observed not only in neurons, vascular endothelial cells, astrocytes, microglia, oligodendrocytes, ependymocytes of diverse cerebral regions, but also in pericytes [60]. Furthermore, neuronal expression of S100a10 even within a single cerebral region was less pronounced, whereas higher expression level was observed in astrocytes and microglia. Moreover, S100a10 expression was significantly increased in the reactive astrocytes upon mouse cerebral damage in the stroke and neuroinflammation models [61].
EAAT-1 and EAAT-2. It was found out that glutamate transporters EAAT-1 (GLAST) [21] and EAAT-2 (GLT-1) [21] were almost exclusively expressed in astrocytes. Moreover, EAAT-1 is the most common marker due to its quite abundant localization in the Bergmann glia, Müller glia, and radial glia [62] (figure). High level of GLT-1 expression was found in the hippocampus, lateral septum, cortex and striatum, whereas GLAST was primarily observed in the cerebellum [21].
It is also worth noting that EAAT-1 (GLAST) and EAAT-2 (GLT-1) display no high specificity. In particular, a synthetic peptide GLT-1 was obtained due to alternative splicing that was primarily expressed in the CNS neurons and peripheral nervous system (PNS), but also found in glial cells (oligodendrocytes, ependymal cells, choroid plexus epithelial cells, satellite cells and Schwann cells) (figure). Moreover, immunostaining with antibodies against the synthetic peptide GLT-1 was mainly detected in the neuronal and glial cytosol allowing to assume that GLT-1 could be present in the intracellular vesicle membranes [20].
Glutamine synthetase (GS). Recently, glutamine synthetase (GS), which is expressed in numerous cerebral regions, has been suggested as another astrocyte marker. GS can bind to diverse astrocyte types including radial glia, Müller glia, and Bergmann glia [22] (figure). It is worth noting that the dual immunofluorescent staining of rat hippocampal slices revealed that the GFAP-positive astroglial cells co-expressed GS, thereby composing a GS-positive astrocyte subpopulation, among which around 40% might contain GFAP-negative cells [63]. In addition, other study with dual immunofluorescent staining of the mouse entorhinal cortex revealed that 10% of the cells co-expressed GS and GFAP, whereas the GFAP-positive cells comprised 12%, but 78% of the glial cells solely expressed GS [64].
In astrocytes, GS is located in the cytosol [65]. Moreover, in vitro study demonstrated that the astrocyte GS could be also found in the vesicle membranes [22].
However, it should be also remembered that the GS expression is observed both in astrocytes as well as in oligodendrocytes, ependymal cells, and some human brain neurons [23] (figure).
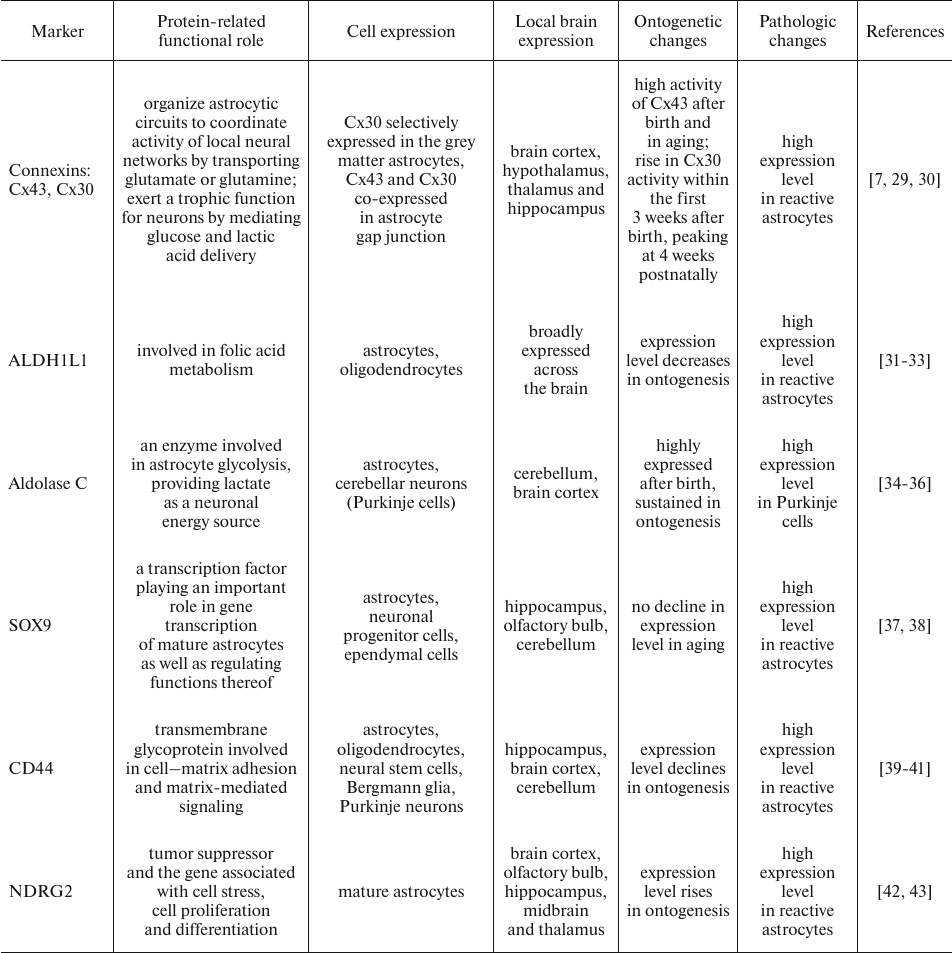
Vimentin. Vimentin is a member of intermediate filament proteins involved in regulating cell migration and differentiation, as well as signal transduction [25]. Vimentin is expressed in astrocytes (figure), and its level could decline gradually in the course of development [66].
It was found out that the mice with ablated GFAP and vimentin-coding genes exhibited impaired formation of the glial scars after the CNS damage as well as altered reactivity in the cerebral vasculature [67]. The latter may be due to the pathological change in endothelial and perivascular cell properties (resulting from the loss of vimentin expression therein) [68] and lowered capacity in the surrounding tissues to withstand blood vessel dilation (due to the lack of GFAP and vimentin in astroglial cells). Altogether, it demonstrates that vimentin might be potentially involved in astrocyte activation resulting from the developed changes in CNS.
Aquaporin 4 (AQP4). Another astrocyte marker is aquaporin 4 (AQP4) belonging to the aquaporin protein family, which form water-transporting channels across cell membranes, also attracts certain interest. This protein is specifically upregulated in the astrocytic endfeet, which are in direct contact with the cerebral microvasculature [27]. In addition, it was also found that to a larger extent AQP4 was enriched solely in the astrocytic endfeet processes [69] (figure).
Connexin (Cx). It is worth mentioning that connexin (Cx) expression in astrocytes is observed across the entire brain being dominated by Cx43 over Cx30, with the latter usually colocalized with Cx43 in the astrocyte gap junctions [70]. Moreover, Cx30 is exclusively expressed in the grey matter astrocytes showing extremely heterogeneous distribution [70, 71]. Connexin expression is necessary for formation of astroglial network able to rapidly transmit some low molecular weight substances (lactate, ATP, NAD+) or ions between cells (figure).
ALDH1L1, Aldolase C, and SOX9. ALDH1L1, which is widely expressed across the entire brain in contrast to the traditional astrocyte marker GFAP, was identified as a new highly-specific astroglial marker in the study [33] investigating the genes expressed in the isolated astrocytes, neurons, and oligodendrocytes. It allowed to reveal a highly branched astrocyte morphology including astrocyte cell body and its processes, whereas GFAP primarily labeled the astrocyte processes (figure). ALDH1L1, also known as 10-formyl-tetrahydrofolate dehydrogenase (FDH) [72], is a folic acid metabolism enzyme converting 10-formyl tetrahydrofolate (10-formyl-THF) into tetrahydrofolate, thus playing a crucial role in multiple reactions such as de novo nucleotide biosynthesis and methionine regeneration, thereby profoundly affecting cell division and growth [31]. Moreover, there is evidence regarding a potential link between the aberrant ALDH1L1 expression and neural tube defects during early CNS development [73]. ALDH1L1 expression changes with age so that ALDH1L1 becomes also expressed in oligodendrocyte subpopulation [32]. Moreover, it was discovered that ALDH1L1 mainly marks cortical astrocytes, but, however, displays a rather weak marking of the white matter astrocytes [74].
Enzyme aldolase C (fructose-1,6-bisphosphate aldolase) is expressed to the greater extent in astrocytes, but the respective gene of aldolase C was also found in the cerebellar neurons (Purkinje cells) [35].
The data obtained during the study investigating expression of the transcription factor SOX9 both in mouse and human brain revealed that SOX9 was almost exclusively expressed in the adult astrocytes except ependymal cells and neurogenic niche, where SOX9 was also detected in the neural precursor cells (figure). It is worth mentioning that SOX9 expression was not reduced with aging, but, on the contrary, was enhanced in the reactive astrocytes in the mouse amyotrophic lateral sclerosis model as well as during the middle cerebral artery occlusion. It should be mentioned that SOX9 is an astrocyte-specific nuclear marker that does not allow to fully reveal astrocyte morphology [37].
In addition, the data from another study [75] demonstrated that CD44 is a plasma membrane protein and extracellular matrix receptor allowing to comprehensively examine astrocyte morphology, which, in turn, could be impossible to do by using GFAP. In particular, the CD44-immunopositive astroglial cells with long processes were discovered in the hippocampus and cortex as well as nearby large blood vessels, and phenotype of such astrocytes was shown to be similar to that of the white matter fibrous astrocytes, but was profoundly different from the protoplasmic astrocyte phenotype. At the same time, the CD44-expressing astroglial cells with short processes displaying phenotype similar to that of both protoplasmic and fibrous astrocytes were also detected in the cortex (figure).
In recent years, NDRG2 (N-myc downstream-regulated gene 2), a tumor suppressor gene related to cell stress, proliferation and differentiation, has been considered as another potential astrocyte marker. It was found out that NDRG2 is expressed in diverse cerebral regions including cortex, olfactory bulb, hippocampus, midbrain, and thalamus, with the maximum expression being observed in the two latter areas [42]. What is most important is that NDRG2 was shown to be selectively expressed in the cerebral astrocytes [42, 76]. Moreover, its expression was upregulated in the glioma cells differentiating to astrocytes, whereas in the reactive astrocytes its level was markedly downmodulated [42]. However, some other studies convincingly demonstrated that the expression of NDRG2 in reactive astrocytes was upregulated in the developing neurodegenerative diseases such as Alzheimer’s and Parkinson’s diseases [43]. NDRG2 could be related to the glial cell proliferation and differentiation and could be considered as a new astrocyte marker especially for the mature non-reactive cerebral astrocytes (figure).
FLUORESCENT DYES AND PROBES
Fluorescent dyes represent a valuable tool for microscopy because they allow to non-invasively investigate neural cell structure and activity without affecting brain function [99].
In particular, astrocytes could be visualized using fluorescent dyes injected into the cells of interest. Usually, several dyes such as Lucifer yellow, Alexa Fluor, biocytin, Fura-2, etc., are used for cell labeling [45, 99]. For instance, the fluorescent dye Fura-2AM (Fura-2-acetoxymethyl ester) is used for measuring intracellular Ca2+ concentration due to its high sensitivity and specificity as well as ability to permeate cell membranes [100].
Alternatively, the Helios Gene Gun transfection system may be used to visualize astrocytes by delivering gold or tungsten particles coated with lipophilic dyes into the live or perfusion fixed brain tissue slices [101]. It allows to rapidly and selectively label cells in various cerebral tissues applying lipophilic dye combinations to facilitate optical separation of closely located cells [102].
Recently, a glyophilic fluorescent probe, cationic dye sulforhodamine 101 and its analogs sulforhodamine B or G, have been suggested for astrocyte visualization [103]. Sulforhodamine undergoes selective uptake by astrocytes and delineates cell structure due to its cytosolic location. Moreover, sulforhodamines could easily penetrate across the blood-brain barrier allowing staining of astrocytes with the intravenously injected sulforhodamine B. Upon that, the astrocyte fluorescence could be observed as early as 40 min post-injection and is sustained for up to 5 h [103]. It seems that in various cerebral regions uptake of sulforhodamine 101 by astrocytes could be mediated by the astrocyte-expressed organic anion transporters. As a result, sulforhodamine 101 efficiently labels hippocampal astrocytes, while it is not accumulated in the astrocytes of the ventrolateral preoptic nucleus [104]. Moreover, it was found out that sulforhodamine 101 selectively labels astrocyte subpopulation of the rat hippocampal slices obtained at early postnatal stage demonstrating typical changes in the course of development as well as traits of the mature GFAP-positive astrocytes [105].
Despite that sulforhodamine 101 efficiently labels astrocytes in the acute rat hippocampal slices [105] as well as mouse cortex in vivo [103], it could also stain oligodendrocytes due to activity of the multi-specific carrier OATP1C1 (SLCO1C1) transporting sulforhodamine 101 from astrocytes via gap junctions of the pan-glial network [106].
Similarly, the coumarin dipeptide, β-Ala-Lys-Nepsilon-Coumarin, specifically labels the rat brain-derived primary astrocytes in culture. However, it also stains oligodendrocyte precursors [107].
Moreover, it is also worth noting that fluorophores such as rhodamines, cyanines, and BODIPYs provide a unique opportunity to selectively and specifically induce strong fluorescence in the mouse and rat hippocampal astrocytes, without labeling neurons, as well as microglia precursor cells or oligodendrocytes [108].
ASTROCYTE-SPECIFIC GENETICALLY ENCODED MARKERS
It is known that astrocytes could be visualized in the cerebral slices by using cell-specific promoter-driven reporter fluorescent proteins selectively expressed in astroglia. For instance, the transgenic mouse strain was created showing the astrocyte-targeted expression of green fluorescent protein (GFP) or its analog enhanced green fluorescent protein (EGFP) driven by GFAP [109] or S100β [110] gene promoter thereby providing a valuable tool for thorough investigation of the dynamic change in the astrocytes during development as well as in pathophysiological states.
It is worth mentioning here that the number of astroglia-specific promoters has been progressively increasing so that by now a set of fluorescent protein probes with distinct spectral characteristics are available [111, 112]. In particular, a series of transgenic mouse strains expressing diverse reef coral fluorescent proteins (RCFP) displaying bright fluorescence covering a wide spectral range (from blue to red) driven by various gene promoters was generated [111].
Moreover, astrocytes could also visualized via expression of the genetically encoded Ca2+ indicators such as Yellow Cameleon-Nano 50 (YC-Nano50) [113] or Green GCaMP consisting of GFP fused to calmodulin and peptide sequence of myosin light chain kinase [114]. Moreover, some studies reported several GCaMP variants including GCaMP2, GCaMP3, GCaMP5, and red-emitting RCaMP tested in astrocytes [115, 116]. In particular, comparing GCaMP2 vs. GCaMP1 after inserting N-terminal 35-mer polyHis RSET (plasmid leader sequence) tag linked to calmodulin and myosin results in the enhanced brightness and thermostability [117]. However, a significant improvement was achieved by constructing GCaMP3 composed of the circularly permuted green fluorescent protein (cpGFP), calmodulin (CaM), and Ca2+/CaM-binding myosin M13 peptide, which displayed increased brightness, wide dynamic range, and higher Ca2+ affinity allowing to report a surge in Ca2+ level triggered by single action potentials in vitro [118]. GCaMP5 was created based on GCaMP3 via site-directed mutagenesis and screening, which allowed to increase dynamic range by severalfold compared to its predecessor GCaMP3 [116]. To generate a red-emitting RCaMP, cpEGFP was replaced with the red fluorescent protein (FP) mRuby in the GCaMP3 scaffold that allowed to increase thermodynamic stability [116].
Remarkably, astrocyte visualization in vivo with fluorescent markers could be also achieved by inoculating adeno-associated viral (AAV) and lentiviral vectors, administration of which could cause emergence of nuclear fluorescent signals specific to the diverse cell types allowing, in turn, to easily identify and distinguish cells of various origin. For instance, by using specific fluorescent labeling of neurons and astrocytes it allowed to distinguish these cell types spectrally due to expression of red fluorescent protein tdTomato in neurons and EGFP in astrocytes [119].
ASSESSING ASTROCYTES GENETIC PROFILE
Level of mRNA characterizes cell transcription state, therefore providing insight into the cell functionality, activity, and development, as well as magnitude of pathological remodeling upon disease progression. Human and mouse brain astrocyte transcriptomic profile were analyzed by using microtemplates and RNA sequencing combined with fluorescence-activated cell sorting (FACS) and cell panning of the astrocytes labeled by the transgene expression [120, 121].
In particular, assessing of the transcriptome in the cortical astroglial cells expressing green fluorescent protein (GFP) and freshly isolated from the adult transgenic FVB/N-Tg (GFAPGFP) 14Mes/J mice by using FACS allowed to demonstrate that the substantial proportion of the astrocyte-expressed genes (around 34%) were involved in cell metabolism [122]. Along with this, the subsequent PCR analysis revealed that the expression level for enzymes taking part in glycogen metabolism was higher in astrocytes than in neurons. While comparing astrocyte vs. neuron it was established that the relative expression of all glycolytic enzymes involved in the reactions starting from glucose phosphorylation to pyruvate synthesis was markedly upregulated in seven out of ten steps of glycolysis. Moreover, astrocytes also exhibited active oxidative metabolism manifested by the relatively high density of mitochondria, which suggested the opportunity for astrocytes to substantially contribute to functional brain visualization including blood-oxygen level-dependent functional magnetic resonance imaging signals [122].
In addition, it was shown using flow cytometry [33] that the S100β/EGFP-positive astrocytes expressing S100β promoter-driven EGFP were present in the S100β-EGFP transgenic mice aged 1-8 and 17-30 days. Comparison of the immature (age 1-8 days old) and mature (age 17-30 days old) astrocytes revealed that not being astrocyte-specific the genes highly expressed in the immature astrocytes are involved in cell proliferation and development (e.g., cell cycle genes), whereas those expressed in the mature astrocytes showed the astroglia-specific pattern. In particular, the latter include the genes encoding secretory proteins such as ApoE, ApoJ/clusterin, Pla2g7, Sparc, Sparcl1, and Mfge, some of which are also important for the astrocyte–synapse or astrocyte–endothelial cell interactions. Many of such genes are highly expressed in the 1-to-17-day-old astrocytes allowing to suggest that the relevant protein products play an important role in the CNS developing during formation of the majority of synapses and afterwards, which, in turn, is in agreement with the data on the ability of astrocytes to regulate synapse formation [123]. Moreover, astrocytes were also found to express other genes (NPAS3, MLC1, LGI1/4, and GPR56) participating in the development of mental diseases such as schizophrenia and bipolar disorder [124-126]. For instance, the NPAS3 gene encoding transcription factor bHLH (basic helix-loop-helix) PAS (Per, Arnt, Sim) domain of the neuronal Protein 3 (NPAS3) is expressed in the Bergmann glia of the cerebellum [127]. The protein NPAS3 takes part in regulating neurogenesis, circadian rhythm, and cell proliferation. NPAS3 gene mutations is considered as a risk factor for mental disorders [128]. The mice with NPAS3 deletion demonstrate development of impaired neurogenesis and synaptic transmission in the hippocampus [129]. The MLC1 gene encoding MLC1 membrane protein is specifically expressed in the distal astroglial processes in the perivascular, subependymal, and subpial areas of the brain. It consists of several transmembrane domains suggesting that it might exert a transport function [124]. The gene LGI1/4 codes for the Leucine-Rich Glioma Inactivated Protein ¼, is expressed in astrocytes, and plays a crucial role in the synaptic transmission and myelination. For instance, LGI4 takes part in regulating proliferation of glial lineage cells in the PNS and promotes Schwann cell myelination via binding to neuronal receptor ADAM22 [130]. The gene GPR56 encodes the G protein-coupled receptor 56 (GPR56), which is expressed both in microglia, oligodendrocytes, and astrocytes, as well as in neuronal precursor cells and developing neurons [131, 132], participates in myelination, proliferation [133], and synaptogenesis [134].
The abovementioned facts allow to extend our understanding of the impact of astrocytes on synaptic regulation and development of mental diseases [135]. Indeed, based on the recently obtained data astrocytes may be directly involved in mediating synapse elimination in the developing and adult brain, which, in turn, plays an important role in remodeling of the brain synapse architecture [136].
The study assessing transcriptome profile of reactive astroglia in astrocytes isolated by flow cytometry from the adult transgenic Aldh1l1-EGFP mice after brain injury modeling seems to be very interesting [61]. It was discovered that the reactive astrocytes profoundly altered their transcriptional profile primarily affecting the genes encoding extracellular matrix proteins such as collagen (Col12a1, Col6a1) and versican (Vcan), thrombospondin (Thbs1) and fibulin 5 (Fbln5), CD44 and neurofascin (Nfasc). It suggests that the reactive astrocytes are able to modify extracellular matrix components during the glial scar formation. Likewise, the genes coding for intermediate filament proteins (GFAP, vimentin and nestin) were highly expressed in the reactive astrocytes, thereby reflecting morphological changes occurring upon activation. Moreover, a gene set encoding cytokines, constituents of the antigen presentation and the complement activation pathway were revealed, which were substantially upregulated in the reactive astrocytes suggesting that astrocytes may exert a regulatory activity upon interaction with immune cells during brain injury [61].
It is important to note that comparing transcriptome profiles among several astrocyte populations expressing specific cell markers, e.g., GFAP- vs. GLT-1-positive astrocytes [122], ALDH1L1- vs. GLT-1-positive astrocytes [32], and ALDH1L1- vs. GlialCAM-positive astrocytes [137] revealed no prominent differences between them suggesting that the majority of astrocyte populations co-expressed such markers.
A single-cell transcriptomic analysis represents one of the tools for determining astrocyte molecular heterogeneity. For instance, several studies investigated the single-cell transcriptomics profile of astrocytes [47, 138], which, however, were not continued to further assess astrocyte heterogeneity. However, a large-scale single-cell RNA sequencing for mouse cortical and hippocampal CA1 cells allowed to identify two astrocyte subsets based on GFAP and Mfge8 expression. In particular, the astrocytes expressing GFAP comprised a glial limiting membrane – the upper layer of the cerebral tissue located immediately beneath the pial membrane, whereas another subset of astrocyte was located more homogenously in the cortex and characterized by less branched processes [47].
OPTOGENETIC ASTROCYTE STIMULATION
Optogenetics represents a valuable tool for investigating astrocyte reaction to transduction of signals specific to the diverse types of neurons based on using opsins as light-sensitive proteins, which include microbial ion channels and ion pumps as well as engineered GPCRs [139]. Opsins absorb certain wavelength of incoming light causing a conformational shift that triggers diverse changes in the opsin-expressing cells. In the process, some opsins [channelrhodopsin-2 (ChR2), halorhodopsin, archerhodopsin (Arch), archerhodopsin-T (Arch-T)] induce ion translocation [140-142], whereas others (melanopsin) activate intracellular cascades such as G-protein signaling pathways [143].
Importantly, optogenetics allows to selectively target certain cell populations particularly astrocytes to be specifically acted upon, which, in turn, allows to conduct deeper and more detailed analysis of their functioning in the CNS [8]. Some studies demonstrated that optogenetic astrocyte stimulation via expression of light-sensitive ion channels and receptors causes significant changes of their functional plasticity [144, 145].
It was found out that stimulation of Opto-a1AR (a metabotropic opsin) expressed in the mouse hippocampal astrocytes could enhance the long-term synaptic plasticity, while the cation membrane channel ChR2 (ionotropic opsin) did not exhibit the so profound effect. It demonstrates that the long-term synaptic plasticity could be modulated optogenetically, which could be used to normalize synaptic transmission and correct impaired plasticity in some neurodegenerative diseases such as Alzheimer’s disease [146].
It was shown that the optically activated astrocyte ChR2 could stimulate ATP release which, in turn, enhanced synaptic excitatory currents via stimulation of P2Y1 and A2A receptors on astrocytes [147]. However, the recent study using the newly designed approach based on the genetically encoded Ca2+ indicator GCaMP6f along with expression of the selective astrocyte melanopsin (G-protein coupled photopigment) to trigger Ca2+ signaling demonstrated that melanopsin was able to stimulate inositol-1,4,5-triphosphate (IP3)-dependent Ca2+ signal transduction and cause the ATP-dependent transient boost of hippocampal excitatory synaptic transmission via purinergic activation of P2Y1 and A2A receptors. Upon that, in contrast to melanopsin, the astrocyte Ca2+ signals induced by ChR2 stimulation were observed after longer (>5 s) light pulses. Moreover, low frequency light stimulation of the melanopsin-transfected astrocytes could cause long-term potentiation that activates episodic-like memory, thereby making melanopsin an optical tool for regulating a wide range of regulatory actions of astrocytes in the neuronal networks in vivo [143].
Another study with the adult (2-5-month-old) tTA-MlC1-tetO-ChR2(C128S)-EYFP mice (developed by Tanaka et al. [148]) investigated sustained Ca2+ response in the optogenetically targeted neocortical astrocytes. In this model, transgenic animals express a highly light-sensitive channelrhodopsin-2 mutant ChR2 (C128S) in astrocytes (under the MlC1 gene promoter), which becomes activated and deactivated with blue-light and yellow-light pulses, therefore allowing to combine optogenetic ChR2 stimulation along with visualized astrocyte Ca2+ activity by using the Ca2+ indicator Rhod-2 AM (red spectral range). Hence, it provided an opportunity to assess astrocyte activation/excitability and determine the role played by astrocytes in pathogenesis of some cerebral diseases such as epilepsy, Alzheimer’s disease, and Parkinson’s disease [149].
Moreover, examining the effects of stimulated astrocytes on input processing in the layer 5 pyramidal neurons (L5PN) revealed that the optogenetically activated astrocytes in the vicinity of L5PN cell body prolong their firing. The observed effects were related to the release of S100β, an astrocytic Ca2+-binding protein decreasing Ca2+ release into extracellular space. Upon that, extracellular Ca2+ level causes L5PN firing mediated by activation of Nav1.6 channels [150].
Interestingly, it was recently discovered that excitotoxicity in the Bergmann glia due to chronic optogenetic ChR2 activation results in the decreased glutamate uptake revealed by increased excitatory postsynaptic currents in Purkinje cells, which is in agreement with the verified downregulated EAAT-1 glutamate transporter expression [151].
Furthermore, physiological role played by astrocytes in vivo was examined in the transgenic Optoα1AR TG mice with astrocytes expressing the optogenetically activated Gq-GPCR Optoα1AR, photostimulation of which ensures a transient rise in the astrocytic Ca2+ level detected by Rhod-2 AM [152]. It allowed to assess the effects of modulated astrocyte stimulation on neuronal activity mediated via the astrocyte Gq signaling. In particular, the optogenetically activated Gq signaling in cortical astrocytes was found to transiently suppress neuronal activity via adenosine A1 receptor. Moreover, sensory stimulation along with the astrocyte activation induced a prolonged depression of the sensory-evoked response. Behavioral testing revealed that the astrocytic activation resulted in the long-term memory enhancement (a novel object recognition task) without affecting the short-term memory (Y-maze) [152].
Considering that the light-induced activation of the astrocyte expressing ChR2 alters the dynamic baseline Ca2+ level [153], mathematical modeling could be a great tool to predict how biophysical properties of the ChR2 construct as well as specifics of laser irradiation may affect astrocyte Ca2+ signaling. In this regard, creating a mathematical model would provide an opportunity to optimize the light stimulation regime to achieve anticipated Ca2+ level in astrocytes as well as to engineer future application-oriented optogenetic variants aimed at comprehensive investigation of the brain astroglial cells [154].
ASTROCYTE PHENOTYPIC ALTERATION IN PATHOPHYSIOLOGY
As a result of negative stimuli contributing to neuronal injury, astrocytes undergo some phenotypic and functional changes accompanied by development of reactive astrogliosis. Upon that, hypertrophy and upregulated GFAP expression are observed that facilitate formation of the typical reactive astrocyte phenotype, which in turn, proliferate, migrate, and are transformed into the scar-forming astrocytes [155]. Similar to the reactive astrocytes, the scar-forming astrocytes demonstrate high expression of a set of protein markers such as GFAP, β-catenin-1, nestin, and N-cadherin. However, there are also gene markers specific for reactive astrocytes – matrix metalloproteinase-2 (MMP-2, encodes MMP-2 protein), PLAUR [soluble urokinase plasminogen activator receptor (suPAR)], matrix metalloproteinase-13 (MMP-13, encodes collagenase 3 protein), Axin-2 (axin-like protein), a gene encoding an intermediate filament protein – nectin, and CTNNB1 (protein catenin β-1), while gene-markers of the scar-forming astrocytes include Cdh2 (N-cadherin), SOX9 (SRY-Box Transcription Factor 9) as well as the chondroitin sulfate proteoglycans-related genes – CSGALNACT1 (chondroitin sulfate N-acetylgalactosaminyltransferase 1), CHST11 (carbohydrate sulfotransferase 11), ACAN (aggrecan), and SLIT2 (Slit Guidance Ligand 2) [155]. Hence, along with the astrocyte morphological characteristics these gene markers may be used for reactive astrocyte and astrocyte phenotyping.
Two types of phenotypes are recognized in the reactive astrocytes – A1 and A2. While the type A1 reactive astrocytes produce pro-inflammatory cytokines able to cause neuronal death, the type A2 astrocytes activate neurotrophic factors contributing to neuronal survival and growth [156]. It must be noted that such reactive astrocyte phenotypes could be identified according to the specific genetic traits. For instance, the post-mortem examination of brain tissues from the patients with Alzheimer’s disease, Huntington’s disease, Parkinson’s disease, amyotrophic lateral sclerosis, and multiple sclerosis revealed that the complement component 3 (C3) was at peak expression in the A1 astrocytes. At the same time, S100A10 was found to be solely expressed in the type A2 astrocytes in the post-mortem brain tissues from multiple sclerosis patients [157]. Hence, it provides an opportunity to use such genes as markers specific to A1 and A2 astrocytes, respectively.
Activated microglia causes transformation of astrocytes to type A1 astrocytes by the released cytokines IL-1α, TNF, and C1q, each of which is important for inducing A1 astrocyte phenotype [157]. In the process, Mfge8 regulates astrocytic A1/A2 conversion by activating PI3K/Akt-axis and suppressing NF-κB-pathway.
In contrast, the type A2 astrocytes exert a neuroprotective effect on nervous system by secreting neurotrophic factors. Expression of the S100a10 gene in A2 astrocytes is necessary for cell proliferation, membrane repair, and inhibition of cell apoptosis [157]. Moreover, the type A2 astrocytes promote expression of the pro-inflammatory cytokine – transforming growth factor-β (TGFβ), involved in synaptogenesis and exerting a neuroprotective activity.
The data from one of the studies investigating several phenotypes of the activated astrocytes and regulatory mechanisms involved are worth mentioning [158]. It was found in this study that the activated astrocytes in vitro and in vivo displayed two distinct functional phenotypes in regard to expression of pro- or anti-inflammatory genes, glial fibrillar acidic protein, as well as neurotoxic or neuroprotective activity. Moreover, it was also demonstrated that the activated astrocytes released protein lipocalin-2 (LCN2) regulating both astrocyte morphological transformation related to reactive astrogliosis as well as migration and apoptosis. Moreover, LCN2 contributed to the pro-inflammatory astrocyte activation, which, however, occurred simultaneously with the inhibition of IL-4/STAT6-axis signaling by participating in the alternative anti-inflammatory activation. Thus, it allows to assume that the astrocyte LCN2 secretory protein acts as an autocrine modulator in astrocyte functional polarization in the developing neuroinflammation that might be aimed at suppressing pro-inflammatory astrocyte activation as well as related neurodegenerative diseases [158, 159].
ASTROCYTES miRNAs AS POTENTIAL TARGETS FOR NEURODEGENERATIVE
DISEASE THERAPY
MicroRNAs are 21-24 nucleotide-long small regulatory RNAs involved in post-transcriptional gene regulation via binding to messenger RNAs (mRNAs) based on complementarity sequence, which results in degradation or suppressed translation of the latter [160]. Moreover, microRNAs possess a potential to control hundreds of diverse transcripts [161]. One of the crucial features related to microRNA is its ability to be transported between the cells that allows microRNAs to exhibit a broad influence at tissue and organismal level [162].
It is known that virtually all cells secrete exosomes [163]. In particular, the recent study [164] investigating molecular profile of astrocytes and neuronal exosomes released into the extracellular space and ensuring intercellular communication via transfer of diverse cues (proteins, RNAs, lipids) showed that only some cellular microRNAs were selectively incorporated into exosomes both in astrocytes and neurons. Furthermore, the existing difference in the microRNA profiles between the exosome and the relevant cell type implies that the process of selective microRNA loading into exosomes is tightly regulated and may show a cell type-specificity.
Both acute and chronic CNS lesions cause reactive, structural, and functional changes in astrocytes defined as reactive astrogliosis. Impaired astrocyte functioning may substantially contribute to the progression of pathological process due to activated excitotoxicity, oxidative stress, and inflammatory responses [165]. Neurodegenerative diseases, Alzheimer’s disease, Parkinson’s disease and amyotrophic lateral sclerosis, in particular, are characterized by neuroinflammation, cognitive and motor dysfunction, which is related to irreversible neuronal degeneration and apoptosis in the brain and spinal cord. However, it has been increasingly evident that the experimental data demonstrate a crucial role played by astrocytes in pathogenesis of chronic neurodegenerative diseases [166].
Impaired regulation of gene expression is a typical trait both during physiological senescence and neurodegenerative diseases [167]. Some studies suggest that microRNAs are involved in pathogenesis of neurodegenerative diseases by regulating astrocyte functions via diverse mechanisms, thereby able to act as potential therapeutic targets. For instance, the mice with the genetically modelled Alzheimer’s disease (3xTg-AD strain) were shown to have elevated level of miR-181a that suppresses expression of certain synthetic proteins in astrocytes, GLT-1 in particular, enabling regulation of synaptic plasticity [168]. Moreover, miR-155 and c-Jun were observed to become activated during the amyloid-beta-induced increase in microglia and astrocyte activation that contributed to production of inflammatory cues such as IL-6 (interleukin-6) and IFN-β (interferon-beta) [169].
It was shown that the mouse model of Alzheimer’s disease (APPswe/PS1/E9 strain) was associated with activated astroglial CEBPD (CCAAT Enhancer Binding Protein Delta – a key transcription factor during cerebral inflammation) that resulted in the altered spatial learning and memory due to activated miR-135a that inhibits activity of the neurotropic factor Thbs1 (thrombospondin 1), which, in turn, suppresses synaptogenesis as well as reduces dendrite growth and axonal sprouting [170].
Parkinson’s disease is associated with diverse pathological factors including α-synuclein toxicity and other mechanisms. It was discovered that the serum levels of miR-153 and miR-223 decreased progressively in the wild type (WT) mice and the genetically engineered Parkinson’s disease model mice (GFAP.HMOX1 strain) [171]. Along with that, the circulatory level of both microRNAs was lower in the transgenic vs. control WT mice, whereas α-synuclein level was elevated in the GFAP.HMOX1 mice vs. the WT mice. Hence, it allowed to suggest that miR-153 and miR-223 negatively regulate α-synuclein in the basal ganglia of the GFAP.HMOX1 mice [171].
On the other hand, it is also worth noting the study [172] assessing survival of the motor neuron exposed to extracellular vesicles secreted by the astrocytes isolated from the patients with amyotrophic lateral sclerosis. It was found that the extracellular vesicles could not only induce death of the motor neurons, but also contained microRNAs that negatively regulate gene expression particularly showing that the miR-494-3p suppresses expression of the genes including the one encoding semaphorin 3A (SEMA3A), which is involved in axonal growth and maintenance.
Noteworthy is the fact that the microRNA isolated from the motor neurons damaged during development of amyotrophic lateral sclerosis could also exert direct negative effect on the structural and functional integrity of astrocytes, which intensified neuronal degeneration significantly [173]. For example, the in vitro experiments data showed that astrocytes were able to take up neuronal miR-218, which, in turn, inhibited activity of the astrocyte glutamate transporter EAAT-2 due to direct impact on 3′-UTR EAAT-2 bearing several sites for miR-218 binding. However, blocking of the miR-218 caused significant increase of the EAAT-2 level in the mouse astrocytes in the amyotrophic lateral sclerosis model, thereby posing it as a promising therapeutic target. Thus, astrocyte dysfunction and reactive astrogliosis occurring due to the motor neuron injury in the developing amyotrophic lateral sclerosis could be mediated by the increased activity of microRNAs released from the damaged neurons [173].
Taken together, both in vivo and in vitro data indicate that microRNAs substantially impact pathogenesis of neurodegenerative diseases by directly or indirectly regulating astrocyte functions, which is manifested by activation of pro-inflammatory cytokines, suppressed synaptogenesis, neuron death, and mitochondrial dysfunction.
CONCLUSIONS
Astrocytes are crucial regulators in the developing and homeostatic brain as well as in emerging and progressing neurodegenerative diseases. Heterogeneity of astrocytes manifested by the diverse brain location-specific morphological traits is an important feature, which could complicate their further investigation. In connection with this, a detailed study of astrocyte gene expression patterns and their functional features in different brain areas may be of paramount importance. Unfortunately, this issue has not been fully resolved yet due to the lack of a universal marker for labeling astroglial cells typically observed in various brain regions. Moreover, none of the ideal astroglia-specific promoter ensuring reporter gene expression solely in this type of cells has been identified yet.
Hence, investigating molecular mechanisms underlying astrocyte–neuron intercellular interaction as well as discovery of novel functions of astroglial cells facilitates the search of new markers of activated astrocytes as well as approaches for their visualization. Combining visualization techniques with modern assays (PCR analysis, electrophysiology, optogenetics) would allow to obtain deeper insights into the multifaceted astrocyte functions, which could be crucial for understanding biology of the central nervous system astroglia in health and disease.
Contributions. Concept (Alla B. Salmina, Olga L. Vlasova), writing the text (Yana V. Gorina), designing drawings (Evgeniy I. Gerasimov), editing the manuscript (Alexander I. Erofeev, Anastasia V. Bolshakova), critical revision for intellectual content (Pavel M. Balaban, Ilya B. Bezprozvanny), approval of the final version of the article for publication (Alla B. Salmina, Olga L. Vlasova).
Funding. The study was financially supported by the Russian Science Foundation (grant no. 20-65-46004).
Ethics declarations. The authors declare no conflicts of interest in financial or any other sphere. This article does not contain any studies with human participants or animals performed by any of the authors.
REFERENCES
1.Montgomery, D. L. (1994) Astrocytes: form,
functions, and roles in disease, Vet. Pathol., 31,
145-167, doi: 10.1177/030098589403100201.
2.Montana, V., Flint, D., Waagepetersen, H. S.,
Schousboe, A., and Parpura, V. (2021) Two metabolic fuels, glucose and
lactate, differentially modulate exocytotic glutamate release from
cultured astrocytes, Neurochem. Res., 46, 2551-2579, doi:
10.1007/s11064-021-03340-y.
3.Kumar, R., Huang, Y.-T., Chen, C.-C., Tzeng, S.-F.,
and Chan, C.-K. (2020) Astrocytic regulation of synchronous bursting in
cortical cultures: from local to global, Cerebral Cortex
Commun., 1, tgaa053, doi: 10.1093/texcom/tgaa053.
4.Honoré, E., Khlaifia, A., Bosson, A., and
Lacaille, J.-C. (2021) Hippocampal somatostatin interneurons, long-term
synaptic plasticity and memory, Front. Neural. Circuits,
15, 687558, doi: 10.3389/fncir.2021.687558.
5.Welser-Alves, J. V., Crocker, S. J., and Milner, R.
(2011) A dual role for microglia in promoting tissue inhibitor of
metalloproteinase (TIMP) expression in glial cells in response to
neuroinflammatory stimuli, J. Neuroinflamm., 8, 61, doi:
10.1186/1742-2094-8-61.
6.Stackhouse, T. L., and Mishra, A. (2021)
Neurovascular coupling in development and disease: focus on astrocytes,
Front. Cell Dev. Biol., 9, 702832, doi:
10.3389/fcell.2021.702832.
7.Santello, M., Toni, N., and Volterra, A. (2019)
Astrocyte function from information processing to cognition and
cognitive impairment, Nat. Neurosci., 22, 154-166, doi:
10.1038/s41593-018-0325-8.
8.McNeill, J., Rudyk, C., Hildebrand, M. E., and
Salmaso, N. (2021) Ion channels and electrophysiological properties of
astrocytes: implications for emergent stimulation technologies,
Front. Cell. Neurosci., 15, 644126, doi:
10.3389/fncel.2021.644126.
9.Gómez-Gonzalo, M., Zehnder, T., Requie, L.
M., Bezz, I. P., and Carmignoto, G. (2018) Insights into the release
mechanism of astrocytic glutamate evoking in neurons NMDA
receptor-mediated slow depolarizing inward currents, Glia,
66, 2188-2199, doi: 10.1002/glia.23473.
10.Robin, L. M., Oliveira da Cruz, J. F., Langlais,
V. C., Martin-Fernandez, M., Metna-Laurent, M., et al. (2018)
Astroglial CB1 receptors determine synaptic D-serine availability to
enable recognition memory, Neuron, 98, 935-944, doi:
10.1016/j.neuron.2018.04.034.
11.Sardar, D., Lozzi, B., Woo, J., Huang, T.-W.,
Cvetkovic, C., et al. (2021) Mapping astrocyte transcriptional
signatures in response to neuroactive compounds, Int. J. Mol.
Sci., 22, 3975, doi: 10.3390/ijms22083975.
12.Gómez-Gonzalo, M., Martin-Fernandez, M.,
Martínez-Murillo, R., Mederos, S., Hernández-Vivanco, A.,
et al. (2017) Neuron-astrocyte signaling is preserved in the aging
brain: neuron-astrocyte signaling in aging brain, Glia,
65, 569-580, doi: 10.1002/glia.23112.
13.Verkhratsky, A., and Nedergaard, M. (2018)
Physiology of astroglia, Physiol. Rev., 98, 239-389, doi:
10.1152/physrev.00042.2016.
14.Middeldorp, J., and Hol, E. M. (2011) GFAP in
health and disease, Progr. Neurobiol., 93, 421-443, doi:
10.1016/j.pneurobio.2011.01.005.
15.Nichols, N. R., Day, J. R., Laping, N. J.,
Johnson, S. A., and Finch, C. E. (1993) GFAP mRNA increases with age in
rat and human brain, Neurobiol. Aging, 14, 421-429, doi:
10.1016/0197-4580(93)90100-P.
16.Garcia, A. D. R., Doan, N. B., Imura, T., Bush,
T. G., and Sofroniew, M. V. (2004) GFAP-expressing progenitors are the
principal source of constitutive neurogenesis in adult mouse forebrain,
Nat. Neurosci., 7, 1233-1241, doi: 10.1038/nn1340.
17.Steiner, J., Bernstein, H.-G., Bogerts, B., Gos,
T., Richter-Landsberg, C., et al. (2008) S100β is expressed in,
and released from, OLN-93 oligodendrocytes: influence of serum and
glucose deprivation, Neuroscience, 154, 496-503, doi:
10.1016/j.neuroscience.2008.03.060.
18.Steiner, J., Bernstein, H.-G., Bielau, H.,
Berndt, A., Brisch, R., et al. (2007) Evidence for a wide
extra-astrocytic distribution of S100β in human brain, BMC
Neurosci., 8, 2, doi: 10.1186/1471-2202-8-2.
19.Marichal, N., García, G., Radmilovich, M.,
Trujillo‐Cenóz, O., and Russo, R. E. (2012) Spatial
domains of progenitor-like cells and functional complexity of a stem
cell niche in the neonatal rat spinal cord, Stem Cells,
30, 2020-2031, doi: 10.1002/stem.1175.
20.Schmitt, A., Asan, E., Lesch, K.-P., and Kugler,
P. (2002) A splice variant of glutamate transporter GLT1/EAAT2
expressed in neurons: cloning and localization in rat nervous system,
Neuroscience, 109, 45-61, doi:
10.1016/S0306-4522(01)00451-1.
21.Lehre, K., Levy, L., Ottersen, O.,
Storm-Mathisen, J., and Danbolt, N. (1995) Differential expression of
two glial glutamate transporters in the rat brain: quantitative and
immunocytochemical observations, J. Neurosci., 15,
1835-1853, doi: 10.1523/JNEUROSCI.15-03-01835.1995.
22.Anlauf, E., and Derouiche, A. (2013) Glutamine
synthetase as an astrocytic marker: its cell type and vesicle
localization, Front. Endocrinol., 4, 144, doi:
10.3389/fendo.2013.00144.
23.Bernstein, H.-G., Bannier, J., Meyer-Lotz, G.,
Steiner, J., Keilhoff, G., et al. (2014) Distribution of immunoreactive
glutamine synthetase in the adult human and mouse brain. Qualitative
and quantitative observations with special emphasis on extra-astroglial
protein localization, J. Chem. Neuroanatomy, 61-62,
33-50, doi: 10.1016/j.jchemneu.2014.07.003.
24.Morrow, C. S., Porter, T. J., Xu, N., Arndt, Z.
P., Ako-Asare, K., et al. (2020) Vimentin coordinates protein turnover
at the aggresome during neural stem Cell quiescence exit, Cell Stem
Cell, 26, 558-568, doi: 10.1016/j.stem.2020.01.018.
25.Battaglia, R. A., Delic, S., Herrmann, H., and
Snider, N. T. (2018) Vimentin on the move: new developments in cell
migration, F1000Res., 7, 1796, doi:
10.12688/f1000research.15967.1.
26.Pattabiraman, S., Azad, G. K., Amen, T., Brielle,
S., Park, J. E., et al. (2020) Vimentin protects differentiating stem
cells from stress, Sci. Rep., 10, 19525, doi:
10.1038/s41598-020-76076-4.
27.Lisjak, M., Potokar, M., Zorec, R., and
Jorgačevski, J. (2020) Indirect role of AQP4b and AQP4d isoforms
in dynamics of astrocyte volume and orthogonal arrays of particles,
Cells, 9, 735, doi: 10.3390/cells9030735.
28.Wen, H., Nagelhus, E. A., Amiry-Moghaddam, M.,
Agre, P., Ottersen, O. P., et al. (1999) Ontogeny of water transport in
rat brain: postnatal expression of the aquaporin-4 water channel:
Aquaporin-4 and brain development, Eur. J. Neurosci., 11,
935-945, doi: 10.1046/j.1460-9568.1999.00502.x.
29.Giaume, C., Naus, C. C., Sáez, J. C., and
Leybaert, L. (2021) Glial connexins and pannexins in the healthy and
diseased brain, Physiol. Rev., 101, 93-145, doi:
10.1152/physrev.00043.2018.
30.Griemsmann, S., Höft, S. P., Bedner, P.,
Zhang, J., von Staden, E., et al. (2015) Characterization of Panglial
gap junction networks in the thalamus, neocortex, and hippocampus
reveals a unique population of glial cells, Cereb. Cortex,
25, 3420-3433, doi: 10.1093/cercor/bhu157.
31.Krupenko, S. A. (2009) FDH: an aldehyde
dehydrogenase fusion enzyme in folate metabolism, Chem. Biol.
Interact., 178, 84-93, doi: 10.1016/j.cbi.2008.09.007.
32.Yang, Y., Vidensky, S., Jin, L., Jie, C.,
Lorenzini, I., et al. (2011) Molecular comparison of GLT1+
and ALDH1L1+ astrocytes in vivo in astroglial
reporter mice, Glia, 59, 200-207, doi:
10.1002/glia.21089.
33.Cahoy, J. D., Emery, B., Kaushal, A., Foo, L. C.,
Zamanian, J. L., et al. (2008) A Transcriptome database for astrocytes,
neurons, and oligodendrocytes: a new resource for understanding brain
development and function, J. Neurosci., 28, 264-278, doi:
10.1523/JNEUROSCI.4178-07.2008.
34.Shiokawa, K., Kajita, E., Hara, H., Yatsuki, H.,
and Hori, K. (2002) A developmental biological study of aldolase gene
expression in Xenopus laevis, Cell Res.,
12, 85-96, doi: 10.1038/sj.cr.7290114.
35.Walther, E. U., Dichgans, M., Maricich, S. M.,
Romito, R. R., Yang, F., et al. (1998) Genomic sequences of aldolase C
(Zebrin II) direct lacZ expression exclusively in non-neuronal cells of
transgenic mice, Proc. Natl. Acad. Sci. USA, 95,
2615-2620, doi: 10.1073/pnas.95.5.2615.
36.Thompson, R. J., Kynoch, P. A. M., and Willson,
V. J. C. (1982) Cellular localization of aldolase C subunits in human
brain, Brain Res., 232, 489-493, doi:
10.1016/0006-8993(82)90294-3.
37.Sun, W., Cornwell, A., Li, J., Peng, S., Osorio,
M. J., et al. (2017) SOX9 is an astrocyte-specific nuclear marker in
the adult brain outside the neurogenic regions, J Neurosci.,
37, 4493-4507, doi: 10.1523/JNEUROSCI.3199-16.2017.
38.Neyrinck, K., Van Den Daele, J., Vervliet, T., De
Smedt, J., Wierda, K., et al. (2021) SOX9-induced generation of
functional astrocytes supporting neuronal maturation in an all-human
system, Stem Cell Rev. Rep., 17, 1855-1873, doi:
10.1007/s12015-021-10179-x.
39.Moretto, G., Xu, R. Y., and Kim, S. U. (1993)
CD44 expression in human astrocytes and oligodendrocytes in culture,
J. Neuropathol. Exp. Neurol., 52, 419-423, doi:
10.1097/00005072-199307000-00009.
40.Dzwonek, J., and Wilczynski, G. M. (2015) CD44:
molecular interactions, signaling and functions in the nervous system,
Front. Cell. Neurosci., 9, 175, doi:
10.3389/fncel.2015.00175.
41.Naruse, M., Shibasaki, K., Yokoyama, S., Kurachi,
M., and Ishizaki, Y. (2013) Dynamic changes of CD44 expression from
progenitors to subpopulations of astrocytes and neurons in developing
cerebellum, PLoS One, 8, e53109, doi:
10.1371/journal.pone.0053109.
42.Shen, L., Zhao, Z.-Y., Wang, Y.-Z., Ji, S.-P.,
Liu, X.-P., et al. (2008) Immunohistochemical detection of Ndrg2 in the
mouse nervous system, NeuroReport., 19, 927-931, doi:
10.1097/WNR.0b013e32830163d0.
43.Tao, L., Zhu, Y., Wang, R., Han, J., Ma, Y., et
al. (2020) N-myc downstream-regulated gene 2 deficiency aggravates
memory impairment in Alzheimer’s disease, Behavioural Brain
Res., 379, 112384, doi: 10.1016/j.bbr.2019.112384.
44.Pekny, M., Eliasson, C., Chien, C.-L., Kindblom,
L. G., Liem, R., et al. (1998) GFAP-deficient astrocytes are capable of
stellation in vitro when cocultured with neurons and exhibit a
reduced amount of intermediate filaments and an increased cell
saturation density, Exp. Cell Res., 239, 332-343, doi:
10.1006/excr.1997.3922.
45.Bushong, E. A., Martone, M. E., Jones, Y. Z., and
Ellisman, M. H. (2002) Protoplasmic astrocytes in CA1 stratum radiatum
occupy separate anatomical domains, J. Neurosci., 22,
183-192, doi: 10.1523/JNEUROSCI.22-01-00183.2002.
46.Van Den Berge, S. A., Middeldorp, J., Zhang, C.
E., Curtis, M. A., Leonard, B. W., et al. (2010) Longterm quiescent
cells in the aged human subventricular neurogenic system specifically
express GFAP-δ: GFAP-δ in aged human SVZ stem cells,
Aging Cell, 9, 313-326, doi:
10.1111/j.1474-9726.2010.00556.x.
47.Zeisel, A., Muñoz-Manchado, A. B.,
Codeluppi, S., Lönnerberg, P., La Manno, G., et al. (2015) Cell
types in the mouse cortex and hippocampus revealed by single-cell
RNA-seq, Science, 347, 1138-1142, doi:
10.1126/science.aaa1934.
48.Hu, J., and Van Eldik, L. J. (1996) S100 β
induces apoptotic cell death in cultured astrocytes via a nitric
oxide-dependent pathway, Biochim. Biophys. Acta Mol. Cell Res.,
1313, 239-245, doi: 10.1016/0167-4889(96)00095-X.
49.Xiong, Z., O’Hanlon, D., Becker, L. E.,
Roder, J., MacDonald, J. F., et al. (2000) Enhanced calcium transients
in glial cells in neonatal cerebellar cultures derived from S100β
null mice, Exp. Cell Res., 257, 281-289, doi:
10.1006/excr.2000.4902.
50.Bianchi, R., Verzini, M., Garbuglia, M.,
Giambanco, I., and Donato, R. (1994) Mechanism of S100
protein-dependent inhibition of glial fibrillary acidic protein (GFAP)
polymerization, Biochim. Biophys. Acta Mol. Cell Res.,
1223, 354-360, doi: 10.1016/0167-4889(94)90095-7.
51.Villarreal, A., Seoane, R., González
Torres, A., Rosciszewski, G., Angelo, M. F., et al. (2014) S100B
protein activates a RAGE-dependent autocrine loop in astrocytes:
implications for its role in the propagation of reactive gliosis, J.
Neurochem., 131, 190-205, doi: 10.1111/jnc.12790.
52.Nishiyama, H., Knöpfel, T., Endo, S., and
Itohara, S. (2002) Glial protein S100B modulates long-term neuronal
synaptic plasticity, Proc. Natl. Acad. Sci. USA, 99,
4037-4042, doi: 10.1073/pnas.052020999.
53.Heizmann, C. W. (2019) S100 proteins: Diagnostic
and prognostic biomarkers in laboratory medicine, Biochim. Biophys.
Acta Mol. Cell Res., 1866, 1197-1206, doi:
10.1016/j.bbamcr.2018.10.015.
54.Ogata, K., and Kosaka, T. (2002) Structural and
quantitative analysis of astrocytes in the mouse hippocampus,
Neuroscience, 113, 221-233, doi:
10.1016/S0306-4522(02)00041-6.
55.Zhang, Z., Ma, Z., Zou, W., Guo, H., Liu, M., et
al. (2019) The appropriate marker for astrocytes: comparing the
distribution and expression of three astrocytic markers in different
mouse cerebral regions, Biomed. Res. Int., 2019,
9605265, doi: 10.1155/2019/9605265.
56.Hachem, S., Aguirre, A., Vives, V., Marks, A.,
Gallo, V., et al. (2005) Spatial and temporal expression of S100B in
cells of oligodendrocyte lineage, Glia, 51, 81-97, doi:
10.1002/glia.20184.
57.Savchenko, V. L., McKanna, J. A., Nikonenko, I.
R., and Skibo, G. G. (2000) Microglia and astrocytes in the adult rat
brain: comparative immunocytochemical analysis demonstrates the
efficacy of lipocortin 1 immunoreactivity, Neuroscience,
96, 195-203, doi: 10.1016/S0306-4522(99)00538-2.
58.Rickmann, M., and Wolff, J. R. (1995) S100
protein expression in subpopulations of neurons of rat brain,
Neuroscience, 67, 977-991, doi:
10.1016/0306-4522(94)00615-C.
59.Zimmer, D. B., Chaplin, J., Baldwin, A., and
Rast, M. (2005) S100‐mediated signal transduction in the nervous
system and neurological diseases, Cell Mol. Biol., 51,
201-214.
60.Milosevic, A., Liebmann, T., Knudsen, M.,
Schintu, N., Svenningsson, P., et al. (2017) Cell- and region-specific
expression of depression-related protein p11 (S100a10) in the brain,
J. Comp. Neurol., 525, 955-975, doi:
10.1002/cne.24113.
61.Zamanian, J. L., Xu, L., Foo, L. C., Nouri, N.,
Zhou, L., et al. (2012) Genomic analysis of reactive astrogliosis,
J. Neurosci., 32, 6391-6410, doi:
10.1523/JNEUROSCI.6221-11.2012.
62.Furuta, A., Rothstein, J. D., and Martin, L. J.
(1997) Glutamate transporter protein subtypes are expressed
differentially during rat CNS development, J. Neurosci.,
17, 8363-8375, doi: 10.1523/JNEUROSCI.17-21-08363.1997.
63.Walz, W., and Lang, M. K. (1998)
Immunocytochemical evidence for a distinct GFAP-negative subpopulation
of astrocytes in the adult rat hippocampus, Neurosci. Lett.,
257, 127-130, doi: 10.1016/S0304-3940(98)00813-1.
64.Yeh, C.-Y., Verkhratsky, A., Terzieva, S., and
Rodríguez, J. J. (2013) Glutamine synthetase in astrocytes from
entorhinal cortex of the triple transgenic animal model of
Alzheimer’s disease is not affected by pathological progression,
Biogerontology, 14, 777-787, doi:
10.1007/s10522-013-9456-1.
65.Norenberg, M. D., and Martinez-Hernandez, A.
(1979) Fine structural localization of glutamine synthetase in
astrocytes of rat brain, Brain Res., 161, 303-310, doi:
10.1016/0006-8993(79)90071-4.
66.Sancho-Tello, M., Vallés, S., Montoliu,
C., Renau-Piqueras, J., and Guerri, C. (1995) Developmental pattern of
GFAP and vimentin gene expression in rat brain and in radial glial
cultures, Glia, 15, 157-166, doi:
10.1002/glia.440150208.
67.Pekny, M., Johansson, C. B., Eliasson, C.,
Stakeberg, J., Wallén, Å., et al. (1999) Abnormal reaction
to central nervous system injury in mice lacking glial fibrillary
acidic protein and vimentin, J. Cell Biol., 145, 503-514,
doi: 10.1083/jcb.145.3.503.
68.Gabbiani, G., Schmid, E., Winter, S., Chaponnier,
C., de Ckhastonay, C., et al. (1981) Vascular smooth muscle cells
differ from other smooth muscle cells: predominance of vimentin
filaments and a specific alpha-type actin, Proc. Natl. Acad. Sci.
USA, 78, 298-302, doi: 10.1073/pnas.78.1.298.
69.Nagelhus, E. A., Veruki, M. L., Torp, R., Haug,
F.-M., Laake, J. H., et al. (1998) Aquaporin-4 water channel protein in
the rat retina and optic nerve: polarized expression in müller
cells and fibrous astrocytes, J. Neurosci., 18,
2506-2519, doi: 10.1523/JNEUROSCI.18-07-02506.1998.
70.Nagy, J. I., Patel, D., Ochalski, P. A. Y., and
Stelmack, G. L. (1999) Connexin30 in rodent, cat and human brain:
selective expression in gray matter astrocytes, co-localization with
connexin43 at gap junctions and late developmental appearance,
Neuroscience, 88, 447-468, doi:
10.1016/S0306-4522(98)00191-2.
71.Theis, M., and Giaume, C. (2012) Connexin-based
intercellular communication and astrocyte heterogeneity, Brain
Res., 1487, 88-98, doi: 10.1016/j.brainres.2012.06.045.
72.Neymeyer, V., Tephly, T. R., and Miller, M. W.
(1997) Folate and 10-formyltetrahydrofolate dehydrogenase (FDH)
expression in the central nervous system of the mature rat, Brain
Res., 766, 195-204, doi: 10.1016/S0006-8993(97)00528-3.
73.Anthony, T. E., and Heintz, N. (2007) The folate
metabolic enzyme ALDH1L1 is restricted to the midline of the early CNS,
suggesting a role in human neural tube defects, J. Comp.
Neurol., 500, 368-383, doi: 10.1002/cne.21179.
74.Waller, R., Woodroofe, M. N., Wharton, S. B.,
Ince, P. G., Francese, S., et al. (2016) Gene expression profiling of
the astrocyte transcriptome in multiple sclerosis normal appearing
white matter reveals a neuroprotective role, J. Neuroimmunol.,
299, 139-146, doi: 10.1016/j.jneuroim.2016.09.010.
75.Sosunov, A. A., Wu, X., Tsankova, N. M.,
Guilfoyle, E., McKhann, G. M., et al. (2014) Phenotypic heterogeneity
and plasticity of isocortical and hippocampal astrocytes in the human
brain, J. Neurosci., 34, 2285-2298, doi:
10.1523/JNEUROSCI.4037-13.2014.
76.Nichols, N. R. (2003) Ndrg2, a novel gene
regulated by adrenal steroids and antidepressants, is highly expressed
in astrocytes, Ann. NY Acad. Sci., 1007, 349-356, doi:
10.1196/annals.1286.034.
77.Docampo-Seara, A., Santos-Durán, G. N.,
Candal, E., Ángel, M., and Díaz, R. (2019) Expression of
radial glial markers (GFAP, BLBP and GS) during telencephalic
development in the catshark (Scyliorhinus canicula),
Brain Struct. Funct., 224, 33-56, doi:
10.1007/s00429-018-1758-2.
78.Shibata, T., Yamada, K., Watanabe, M., Ikenaka,
K., Wada, K., et al. (1997) Glutamate transporter GLAST is expressed in
the radial glia-astrocyte lineage of developing mouse spinal cord,
J. Neurosci., 17, 92129-219, doi:
10.1523/JNEUROSCI.17-23-09212.1997.
79.Belozor, O. S., Yakovleva, D. A., Potapenko, I.
V., Shuvaev, A. N., Smolnikova, M. V., et al. (2019) Extracellular S100
disrupts Bergman glia morphology and synaptic transmission in
cerebellar Purkinje cells, Brain Sci., 9, 80, doi:
10.3390/brainsci9040080.
80.Miyazaki, T., Yamasaki, M., Hashimoto, K., Kohda,
K., Yuzaki, M., et al. (2017) Glutamate transporter GLAST controls
synaptic wrapping by Bergmann glia and ensures proper wiring of
Purkinje cells, Proc. Natl. Acad. Sci. USA, 114,
7438-7443, doi: 10.1073/pnas.1617330114.
81.Tiburcio-Félix, R.,
Escalante-López, M., López-Bayghen, B., Martínez,
D., Hernández-Kelly, L. C., et al. (2018) Glutamate-dependent
translational control of glutamine synthetase in bergmann glia cells,
Mol. Neurobiol., 55, 5202-5209, doi:
10.1007/s12035-017-0756-3.
82.Kommata, V., and Dermon, C. R. (2018) Transient
vimentin expression during the embryonic development of the chicken
cerebellum, Int. J. Dev. Neurosci., 65, 11-20, doi:
10.1016/j.ijdevneu.2017.10.003.
83.Pow, D. V., and Barnett, N. L. (1999) Changing
patterns of spatial buffering of glutamate in developing rat retinae
are mediated by the Müller cell glutamate transporter GLAST,
Cell Tissue Res., 297, 57-66, doi:
10.1007/s004410051333.
84.Yu, J., Zhong, Y., Cheng, Y., Shen, X., Wang, J.,
et al. (2011) Effect of high hydrostatic pressure on the expression of
glutamine synthetase in rat retinal Müller cells cultured in
vitro, Exp. Ther. Med., 2, 513-516, doi:
10.3892/etm.2011.239.
85.Voulgaris, D., Nikolakopoulou, P., and Herland,
A. (2022) Generation of human iPSC-derived astrocytes with a mature
star-shaped phenotype for CNS modeling, Stem Cell Rev. Rep.,
doi: 10.1007/s12015-022-10376-2.
86.Verkhratsky, A., Marutle, A.,
Rodríguez-Arellano, J. J., and Nordberg, A. (2015) Glial
asthenia and functional paralysis: a new perspective on
neurodegeneration and Alzheimer’s disease, Neuroscientist,
21, 552-568, doi: 10.1177/1073858414547132.
87.Choi, S. R., Roh, D. H., Yoon, S. Y., Kwon, S.
G., Choi, H. S., et al. (2016) Astrocyte sigma-1 receptors modulate
connexin 43 expression leading to the induction of below-level
mechanical allodynia in spinal cord injured mice,
Neuropharmacology, 111, 34-46, doi:
10.1016/j.neuropharm.2016.08.027.
88.Çavdar, S., Köse, B., Sur-Erdem,
İ., and Özkan, M. (2021) Comparing astrocytic gap junction
of genetic absence epileptic rats with control rats: an experimental
study, Brain Struct. Funct., 226, 2113-2123, doi:
10.1007/s00429-021-02310-y.
89.Sandoval, M., Luarte, A., Herrera-Molina, R.,
Varas-Godoy, M., Santibáñez, M., et al. (2013) The
glycolytic enzyme aldolase C is up-regulated in rat forebrain
microsomes and in the cerebrospinal fluid after repetitive fluoxetine
treatment, Brain Res., 1520, 1-14, doi:
10.1016/j.brainres.2013.04.049.
90.Flügge, G., Araya-Callis, C.,
Garea-Rodriguez, E., Stadelmann-Nessler, C., and Fuchs, E. (2014) NDRG2
as a marker protein for brain astrocytes, Cell Tissue Res.,
357, 31-41, doi: 10.1007/s00441-014-1837-5.
91.Desilva, T. M., Billiards, S. S., Borenstein, N.
S., Trachtenberg, F. L., Volpe, J. J., et al. (2008) Glutamate
transporter EAAT2 expression is up-regulated in reactive astrocytes in
human periventricular leukomalacia, J. Comp. Neurol.,
508, 238-248, doi: 10.1002/cne.21667.
92.Xin, W., Mironova, Y. A., Shen, H., Marino, R. A.
M., Waisman, A., et al. (2019) Oligodendrocytes support neuronal
glutamatergic transmission via expression of glutamine synthetase,
Cell Rep., 27, 2262-2271.e5, doi:
10.1016/j.celrep.2019.04.094.
93.Winchenbach, J., Düking, T., Berghoff, S.
A., Stumpf, S. K., Hülsmann, S., et al. (2016) Inducible targeting
of CNS astrocytes in Aldh1l1-CreERT2 BAC transgenic mice,
F1000Res., 5, 2934, doi:
10.12688/f1000research.10509.1.
94.Tuohy, T. M. F., Wallingford, N., Liu, Y., Chan,
F. H., Rizvi, T., et al. (2004) CD44 overexpression by
oligodendrocytes: a novel mouse model of inflammation-independent
demyelination and dysmyelination, Glia, 47, 335-345, doi:
10.1002/glia.20042.
95.Porlan, E., Martí-Prado, B.,
Morante-Redolat, J. M., Consiglio, A., Delgado, A. C., et al. (2014)
MT5-MMP regulates adult neural stem cell functional quiescence through
the cleavage of N-cadherin, Nat. Cell. Biol., 16,
629-638, doi: 10.1038/ncb2993.
96.Akanuma, S., Sakurai, T., Tachikawa, M., Kubo,
Y., and Hosoya, K. (2015) Transporter-mediated L-glutamate elimination
from cerebrospinal fluid: possible involvement of excitatory amino acid
transporters expressed in ependymal cells and choroid plexus epithelial
cells, Fluids Barriers CNS, 12, 11, doi:
10.1186/s12987-015-0006-x.
97.Trillo-Contreras, J. L., Toledo-Aral, J. J.,
Echevarría, M., and Villadiego, J. (2019) AQP1 and AQP4
contribution to cerebrospinal fluid homeostasis, Cells,
8, 197, doi: 10.3390/cells8020197.
98.Ren, Y., Ao, Y., O’Shea, T. M., Burda, J.
E., Bernstein, A. M., et al. (2017) Ependymal cell contribution to scar
formation after spinal cord injury is minimal, local and dependent on
direct ependymal injury, Sci. Rep., 7, 41122, doi:
10.1038/srep41122.
99.Garaschuk, O., Milos, R. I., and Konnerth, A.
(2006) Targeted bulk-loading of fluorescent indicators for two-photon
brain imaging in vivo, Nat. Protoc., 1, 380-386,
doi: 10.1038/nprot.2006.58.
100.Hideharu, H., and Haruo, M. (1994) Fluorescence
imaging of intracellular Ca2+, J. Pharmacol. Toxicol.
Methods, 31, 1-10, doi: 10.1016/1056-8719(94)90023-X.
101.Benediktsson, A. M., Schachtele, S. J., Green,
S. H., and Dailey, M. E. (2005) Ballistic labeling and dynamic imaging
of astrocytes in organotypic hippocampal slice cultures, J.
Neurosci. Methods, 141, 41-53, doi:
10.1016/j.jneumeth.2004.05.013.
102.Gan, W.-B., Grutzendler, J., Wong, W. T., Wong,
R. O. L., and Lichtman, J. W. (2000) Multicolor “DiOlistic”
labeling of the nervous system using lipophilic dye combinations,
Neuron, 27, 219-225, doi:
10.1016/S0896-6273(00)00031-3.
103.Nimmerjahn, A., and Helmchen, F. (2012) In
vivo labeling of cortical astrocytes with sulforhodamine 101
(SR101), Cold Spring Harb. Protoc., 2012, 326-334, doi:
10.1101/pdb.prot068155.
104.Schnell, C., Hagos, Y., and Hülsmann, S.
(2012) Active sulforhodamine 101 uptake into hippocampal astrocytes,
PLoS One, 7, e49398, doi:
10.1371/journal.pone.0049398.
105.Kafitz, K. W., Meier, S. D., Stephan, J., and
Rose, C. R. (2008) Developmental profile and properties of
sulforhodamine 101 – labeled glial cells in acute brain slices of
rat hippocampus, J. Neurosci. Methods, 169, 84-92, doi:
10.1016/j.jneumeth.2007.11.022.
106.Hagos, L., and Hülsmann, S. (2016)
Unspecific labelling of oligodendrocytes by sulforhodamine 101 depends
on astrocytic uptake via the thyroid hormone transporter OATP1C1
(SLCO1C1), Neurosci. Lett., 631, 13-18, doi:
10.1016/j.neulet.2016.08.010.
107.Dieck, S. T., Heuer, H., Ehrchen, J., Otto, C.,
and Bauer, K. (1999) The peptide transporter PepT2 is expressed in rat
brain and mediates the accumulation of the fluorescent dipeptide
derivative beta-Ala-Lys-Nepsilon-AMCA in astrocytes, Glia,
25, 10-20, doi:
10.1002/(sici)1098-1136(19990101)25:1<10::aid-glia2>3.0.co;2-y.
108.Preston, A. N., Farr, J. D., O’Neill, B.
K., Thompson, K. K., Tsirka, S. E., et al. (2018) Visualizing the
brain’s astrocytes with diverse chemical scaffolds, ACS Chem.
Biol., 13, 1493-1498, doi: 10.1021/acschembio.8b00391.
109.Suzuki, R., Watanabe, J., Arata, S., Funahashi,
H., Kikuyama, S., et al. (2003) A transgenic mouse model for the
detailed morphological study of astrocytes, Neurosci. Res.,
47, 451-454, doi: 10.1016/j.neures.2003.08.008.
110.Vives, V., Alonso, G., Solal, A. C., Joubert,
D., and Legraverend, C. (2003) Visualization of S100B-positive neurons
and glia in the central nervous system of EGFP transgenic mice, J.
Comp. Neurol., 457, 404-419, doi: 10.1002/cne.10552.
111.Hirrlinger, P. G., Scheller, A., Braun, C.,
Quintela-Schneider, M., Fuss, B., et al. (2005) Expression of reef
coral fluorescent proteins in the central nervous system of transgenic
mice, Mol. Cell. Neurosci., 30, 291-303, doi:
10.1016/j.mcn.2005.08.011.
112.Nowotschin, S., Eakin, G. S., and
Hadjantonakis, A.-K. (2009) Live-imaging fluorescent proteins in mouse
embryos: multi-dimensional, multi-spectral perspectives, Trends
Biotechnol., 27, 266-276, doi:
10.1016/j.tibtech.2009.02.006.
113.Kanemaru, K. (2016) Visualization of astrocytic
calcium signals in fine process using an ultrasensitive calcium
indicator YC-Nano50, Folia Pharmacol. Japonica, 147,
190-193, doi: 10.1254/fpj.147.190.
114.Miyawaki, A., Llopis, J., Heim, R., McCaffery,
J. M., Adams, J. A., et al. (1997) Fluorescent indicators for
Ca2+ based on green fluorescent proteins and calmodulin,
Nature, 388, 882-887, doi: 10.1038/42264.
115.Akerboom, J., Chen, T.-W., Wardill, T. J.,
Tian, L., Marvin, J. S., et al. (2012) Optimization of a GCaMP calcium
indicator for neural activity imaging, J. Neurosci., 32,
13819-13840, doi: 10.1523/JNEUROSCI.2601-12.2012.
116.Akerboom, J., Carreras Calderón, N.,
Tian, L., Wabnig, S., Prigge, M., et al. (2013) Genetically encoded
calcium indicators for multi-color neural activity imaging and
combination with optogenetics, Front. Mol. Neurosci., 6,
2, doi: 10.3389/fnmol.2013.00002.
117.Tallin, Y. N. (2006) Imaging cellular signals
in the heart in vivo: cardiac expression of the high-signal
Ca2+ indicator GCaMP2, Proc. Natl. Acad. Sci. USA,
103, 4753-4758, doi: 10.1073/pnas.0509378103.
118.Tian, L., Hires, S. A., Mao, T., Huber, D.,
Chiappe, M. E., et al. (2009) Imaging neural activity in worms, flies
and mice with improved GCaMP calcium indicators, Nat. Methods,
6, 875-881, doi: 10.1038/nmeth.1398.
119.Schulze, W., Hayata-Takano, A., Kamo, T.,
Nakazawa, T., Nagayasu, K., et al. (2015) Simultaneous neuron- and
astrocyte-specific fluorescent marking, Biochem. Biophys. Res.
Commun., 459, 81-86, doi: 10.1016/j.bbrc.2015.02.073.
120.John Lin, C.-C., Yu, K., Hatcher, A., Huang,
T.-W., Lee, H. K., et al. (2017) Identification of diverse astrocyte
populations and their malignant analogs, Nat. Neurosci.,
20, 396-405, doi: 10.1038/nn.4493.
121.Batiuk, M. Y., Martirosyan, A., Wahis, J., de
Vin, F., Marneffe, C., et al. (2020) Identification of region-specific
astrocyte subtypes at single cell resolution, Nat. Commun.,
11, 1220, doi: 10.1038/s41467-019-14198-8.
122.Lovatt, D., Sonnewald, U., Waagepetersen, H.
S., Schousboe, A., He, W., et al. (2007) The transcriptome and
metabolic gene signature of protoplasmic astrocytes in the adult murine
cortex, J. Neuroscience, 27, 12255-12266, doi:
10.1523/JNEUROSCI.3404-07.2007.
123.Christopherson, K. S., Ullian, E. M., Stokes,
C. C. A., Mullowney, C. E., Hell, J. W., et al. (2005) Thrombospondins
are astrocyte-secreted proteins that promote CNS synaptogenesis,
Cell, 120, 421-433, doi: 10.1016/j.cell.2004.12.020.
124.Ilja Boor, P. K., de Groot, K., Waisfisz, Q.,
Kamphorst, W., Oudejans, C. B. M., et al. (2005) MLC1: a novel protein
in distal astroglial processes, J. Neuropathol. Exp. Neurol.,
64, 412-419, doi: 10.1093/jnen/64.5.412.
125.Kinboshi, M., Shimizu, S., Mashimo, T.,
Serikawa, T., Ito, H., et al. (2019) Down-regulation of astrocytic
Kir4.1 Channels during the audiogenic epileptogenesis in leucine-rich
glioma-inactivated 1 (Lgi1) mutant rats, IJMS, 20, 1013,
doi: 10.3390/ijms20051013.
126.Chiou, B., Gao, C., Giera, S., Folts, C. J.,
Kishore, P., et al. (2021) Cell type‐specific evaluation of
ADGRG1/GPR56 function in developmental central nervous system
myelination, Glia, 69, 413-423, doi:
10.1002/glia.23906.
127.Gould, P., and Kamnasaran, D. (2011)
Immunohistochemical analyses of NPAS3 expression in the developing
human fetal brain, Anat. Histol. Embryol., 40, 196-203,
doi: 10.1111/j.1439-0264.2010.01059.x.
128.Kamnasaran, D., Muir, W. J., Ferguson-Smith, M.
A., and Cox, D. W. (2003) Disruption of the neuronal PAS3 gene in a
family affected with schizophrenia, J. Med. Genet., 40,
325-332, doi: 10.1136/jmg.40.5.325.
129.Pieper, A. A., Wu, X., Han, T. W., Estill, S.
J., Dang, Q., et al. (2005) The neuronal PAS domain protein 3
transcription factor controls FGF-mediated adult hippocampal
neurogenesis in mice, Proc. Natl. Acad. Sci. USA,
102, 14052-14057, doi: 10.1073/pnas.0506713102.
130.Linde, K., Aunin, E., Meijer, D., and
Bermingham, J. R. (2013) LGI proteins in the nervous system, ASN
Neuro, 5, 167-181, doi: 10.1042/AN20120095.
131.Bai, Y., Du, L., Shen, L., Zhang, Y., and
Zhang, L. (2009) GPR56 is highly expressed in neural stem cells but
downregulated during differentiation, Neuroreport, 20,
918-922, doi: 10.1097/WNR.0b013e32832c92d7.
132.Zhang, Y., Chen, K., Sloan, S. A., Bennett, M.
L., Scholze, A. R., et al. (2014) An RNA-sequencing transcriptome and
splicing database of glia, neurons, and vascular cells of the cerebral
cortex, J. Neurosci., 34, 11929-11947, doi:
10.1523/JNEUROSCI.1860-14.2014.
133.Giera, S., Luo, R., Ying, Y., Ackerman, S. D.,
Jeong, S. J., et al. (2018) Microglial transglutaminase-2 drives
myelination and myelin repair via GPR56/ADGRG1 in oligodendrocyte
precursor cells, ELife, 7, e33385, doi:
10.7554/eLife.33385.
134.Li, T., Chiou, B., Gilman, C. K., Luo, R.,
Koshi, T., et al. (2020) A splicing isoform of GPR56 mediates
microglial synaptic refinement via phosphatidylserine binding, EMBO
J., 39, e104136, doi: 10.15252/embj.2019104136.
135.Chung, W.-S., Welsh, C. A., Barres, B. A., and
Stevens, B. (2015) Do glia drive synaptic and cognitive impairment in
disease? Nat. Neurosci., 18, 1539-1545, doi:
10.1038/nn.4142.
136.Chung, W.-S., Clarke, L. E., Wang, G. X.,
Stafford, B. K., Sher, A., et al. (2013) Astrocytes mediate synapse
elimination through MEGF10 and MERTK pathways, Nature,
504, 394-400, doi: 10.1038/nature12776.
137.Zhang, Y., Sloan, S. A., Clarke, L. E., Caneda,
C., Plaza, C. A., et al. (2016) Purification and characterization of
progenitor and mature human astrocytes reveals transcriptional and
functional differences with mouse, Neuron, 89, 37-53,
doi: 10.1016/j.neuron.2015.11.013.
138.Tasic, B., Menon, V., Nguyen, T. N., Kim, T.
K., Jarsky, T., et al. (2016) Adult mouse cortical cell taxonomy
revealed by single cell transcriptomics, Nat. Neurosci.,
19, 335-346, doi: 10.1038/nn.4216.
139.Bang, J., Kim, H. Y., and Lee, H. (2016)
Optogenetic and chemogenetic approaches for studying astrocytes and
gliotransmitters, Exp. Neurobiol., 25, 205-221, doi:
10.5607/en.2016.25.5.205.
140.Nagel, G., Szellas, T., Huhn, W., Kateriya, S.,
Adeishvili, N., et al. (2003) Channelrhodopsin-2, a directly
light-gated cation-selective membrane channel, Proc. Natl. Acad.
Sci. USA, 100, 13940-13945, doi:
10.1073/pnas.1936192100.
141.Bekar, L. K., and Walz, W. (1999) Evidence for
chloride ions as intracellular messenger substances in astrocytes,
J. Neurophysiol., 82, 248-254, doi:
10.1152/jn.1999.82.1.248.
142.El-Gaby, M., Zhang, Y., Wolf, K., Schwiening,
C. J., Paulsen, O., et al. (2016) Archaerhodopsin selectively and
reversibly silences synaptic transmission through altered pH, Cell
Rep., 16, 2259, doi: 10.1016/j.celrep.2016.07.057.
143.Mederos, S., Hernández-Vivanco, A.,
Ramírez-Franco, J., Martín-Fernández, M.,
Navarrete, M., et al. (2019) Melanopsin for precise optogenetic
activation of astrocyte-neuron networks, Glia, 67,
915-934, doi: 10.1002/glia.23580.
144.Van Den Herrewegen, Y., Sanderson, T. M., Sahu,
S., De Bundel, D., Bortolotto, Z. A., et al. (2021) Side-by-side
comparison of the effects of Gq- and Gi-DREADD-mediated astrocyte
modulation on intracellular calcium dynamics and synaptic plasticity in
the hippocampal CA1, Mol Brain., 14, 144, doi:
10.1186/s13041-021-00856-w.
145.Nam, M. H., Won, W., Han, K. S., and Lee, C. J.
(2021) Signaling mechanisms of µ-opioid receptor (MOR) in the
hippocampus: disinhibition versus astrocytic glutamate regulation,
Cell. Mol. Life Sci., 78, 415-426, doi:
10.1007/s00018-020-03595-8.
146.Gerasimov, E., Erofeev, A., Borodinova, A.,
Bolshakova, A., Balaban, P., et al. (2021) Optogenetic activation of
astrocytes-effects on neuronal network function, Int. J. Mol.
Sci., 22, 9613, doi: 10.3390/ijms22179613.
147.Shen, W., Nikolic, L., Meunier, C., Pfrieger,
F., and Audinat, E. (2017) An autocrine purinergic signaling controls
astrocyte-induced neuronal excitation, Sci Rep., 7,
11280, doi: 10.1038/s41598-017-11793-x.
148.Tanaka, K. F., Matsui, K., Sasaki, T., Sano,
H., Sugio, S., et al. (2012) Expanding the repertoire of
optogenetically targeted cells with an enhanced gene expression system,
Cell Rep., 2, 397-406, doi:
10.1016/j.celrep.2012.06.011.
149.Balachandar, L., Montejo, K. A., Castano, E.,
Perez, M., Moncion, C., et al. (2020) Simultaneous Ca2+
imaging and optogenetic stimulation of cortical astrocytes in adult
murine brain slices, Curr. Protoc. Neurosci., 94, e110,
doi: 10.1002/cpns.110.
150.Ryczko, D., Hanini-Daoud, M., Condamine, S.,
Bréant, B. J. B., Fougère, M., et al. (2021)
S100β-mediated astroglial control of firing and input processing
in layer 5 pyramidal neurons of the mouse visual cortex, J.
Physiol., 599, 677-707, doi: 10.1113/JP280501.
151.Shuvaev, A. N., Belozor, O. S., Mozhei, O.,
Yakovleva, D. A., Potapenko, I. V., et al. (2021) Chronic optogenetic
stimulation of Bergman glia leads to dysfunction of EAAT1 and Purkinje
cell death, mimicking the events caused by expression of pathogenic
ataxin-1, Neurobiol. Dis., 154, 105340, doi:
10.1016/j.nbd.2021.105340.
152.Iwai, Y., Ozawa, K., Yahagi, K., Mishima, T.,
Akther, S., et al. (2021) Transient astrocytic Gq signaling underlies
remote memory enhancement, Front. Neural Circuits, 15,
658343, doi: 10.3389/fncir.2021.658343.
153.Takata, N., Sugiura, Y., Yoshida, K., Koizumi,
M., Hiroshi, N., et al. (2018) Optogenetic astrocyte activation evokes
BOLD fMRI response with oxygen consumption without neuronal activity
modulation, Glia, 66, 2013-2023, doi:
10.1002/glia.23454.
154.Moshkforoush, A., Lakshmini, B., Moncion, C.,
Montejo, K. A., and Riera, J. (2021) Unraveling ChR2-driven stochastic
Ca2+ dynamics in astrocytes: a call for new interventional
paradigms, PLoS Comput. Biol., 17, e1008648, doi:
10.1371/journal.pcbi.1008648.
155.Hara, M., Kobayakawa, K., Ohkawa, Y., Kumamaru,
H., Yokota, K., et al. (2017) Interaction of reactive astrocytes with
type I collagen induces astrocytic scar formation through the
integrin-N-cadherin pathway after spinal cord injury, Nat. Med.,
23, 818-828, doi: 10.1038/nm.4354.
156.Li, T., Chen, X., Zhang, C., Zhang, Y., and
Yao, W. (2019) An update on reactive astrocytes in chronic pain, J.
Neuroinflammation, 16, 140, doi:
10.1186/s12974-019-1524-2.
157.Liddelow, S. A., Guttenplan, K. A., Clarke, L.
E., Bennett, F. C., Bohlen, C. J., et al. (2017) Neurotoxic reactive
astrocytes are induced by activated microglia, Nature,
541, 481-487, doi: 10.1038/nature21029.
158.Jang, E., Kim, J.-H., Lee, S., Kim, J.-H., Seo,
J.-W., et al. (2013) Phenotypic Polarization of activated astrocytes:
the critical role of lipocalin-2 in the classical inflammatory
activation of astrocytes, J. Immunol., 191, 5204-5219,
doi: 10.4049/jimmunol.1301637.
159.Lee, S., Jha, M. K., and Suk, K. (2015)
Lipocalin-2 in THE Inflammatory activation of brain astrocytes,
Crit. Rev. Immunol., 35, 77-84, doi:
10.1615/CritRevImmunol.2015012127.
160.Jonas, S., and Izaurralde, E. (2015) Towards a
molecular understanding of microRNA-mediated gene silencing, Nat.
Rev. Genet., 16, 421-433, doi: 10.1038/nrg3965.
161.Bartel, D. P., and Chen, C.-Z. (2004)
Micromanagers of gene expression: the potentially widespread influence
of metazoan microRNAs, Nat. Rev. Genet., 5, 396-400, doi:
10.1038/nrg1328.
162.Thakur, B. K., Zhang, H., Becker, A., Matei,
I., Huang, Y., et al. (2014) Double-stranded DNA in exosomes: a novel
biomarker in cancer detection, Cell Res., 24, 766-769,
doi: 10.1038/cr.2014.44.
163.Frühbeis, C., Fröhlich, D., Kuo, W.
P., Amphornrat, J., Thilemann, S., et al. (2013)
Neurotransmitter-triggered transfer of exosomes mediates
oligodendrocyte-neuron communication, PLoS Biol., 11,
e1001604, doi: 10.1371/journal.pbio.1001604.
164.Luo, X., Jean-Toussaint, R., Sacan, A., and
Ajit, S. K. (2021) Differential RNA packaging into small extracellular
vesicles by neurons and astrocytes, Cell. Commun. Signal.,
19, 75, doi: 10.1186/s12964-021-00757-4.
165.Escartin, C., Galea, E., Lakatos, A.,
O’Callaghan, J. P., Petzold, G. C., et al. (2021) Reactive
astrocyte nomenclature, definitions, and future directions, Nat.
Neurosci., 24, 312-325, doi: 10.1038/s41593-020-00783-4.
166.Pekny, M., and Pekna, M. (2016) Reactive
gliosis in the pathogenesis of CNS diseases, Biochim. Biophys.
Acta, 1862, 483-491, doi: 10.1016/j.bbadis.2015.11.014.
167.Gangisetty, O., Cabrera, M. A., and Murugan, S.
(2018) Impact of epigenetics in aging and age related neurodegenerative
diseases, Front. Biosci. (Landmark Ed), 23, 1445-1464,
doi: 10.2741/4654.
168.Zumkehr, J., Rodriguez-Ortiz, C. J., Medeiros,
R., and Kitazawa, M. (2018) Inflammatory cytokine, IL-1β,
regulates glial glutamate transporter via microRNA-181a in
vitro, J. Alzheimer’s Dis., 63, 965-975, doi:
10.3233/JAD-170828.
169.Guedes, J. R., Custódia, C. M., Silva,
R. J., de Almeida, L. P., Pedroso de Lima, M. C., et al. (2014) Early
miR-155 upregulation contributes to neuroinflammation in
Alzheimer’s disease triple transgenic mouse model, Hum. Mol.
Genet., 23, 6286-6301, doi: 10.1093/hmg/ddu348.
170.Chu, Y. Y., Ko, C. Y., Wang, W. J., Wang, S.
M., Gean, P. W., et al. (2016) Astrocytic CCAAT/enhancer binding
protein δ regulates neuronal viability and spatial learning
ability via miR-135a, Mol. Neurobiol., 53, 4173-4188,
doi: 10.1007/s12035-015-9359-z.
171.Cressatti, M., Song, W., Turk, A. Z., Garabed,
L. R., Benchaya, J. A., et al. (2019) Glial HMOX1 expression
promotes central and peripheral α-synuclein dysregulation and
pathogenicity in parkinsonian mice, Glia, 67, 1730-1744,
doi: 10.1002/glia.23645.
172.Varcianna, A., Myszczynska, M. A., Castelli, L.
M., O’Neill, B., Kim, Y., et al. (2019) Micro-RNAs secreted
through astrocyte-derived extracellular vesicles cause neuronal network
degeneration in C9orf72 ALS, EBioMedicine, 40, 626-635,
doi: 10.1016/j.ebiom.2018.11.067.
173.Hoye, M. L., Regan, M. R., Jensen, L. A., Lake,
A. M., Reddy, L. V., et al. (2018) Motor neuron-derived microRNAs cause
astrocyte dysfunction in amyotrophic lateral sclerosis, Brain,
141, 2561-2575, doi: 10.1093/brain/awy182.