The Role of C1473G Polymorphism in Mouse Triptophan Hydroxylase 2 Gene in the Acute Effects of Ethanol on the c-fos Gene Expression and Metabolism of Biogenic Amines in the Brain
Darya V. Bazovkina1,a*, Dariya V. Fursenko1, Vladimir S. Naumenko1, and Aleksandr V. Kulikov1
1Institute of Cytology and Genetics, Siberian Branch of Russian Academy of Sciences, 630090 Novosibirsk, Russia* To whom correspondence should be addressed.
Received November 3, 2022; Revised November 26, 2022; Accepted November 29, 2022
Tryptophan hydroxylase 2 is a key enzyme in the synthesis of the neurotransmitter serotonin, which plays an important role in the regulation of behavior and various physiological functions. We studied the effect of acute ethanol administration on the expression of the early response c-fos gene and metabolism of serotonin and catecholamines in the brain structures of B6-1473C and B6-1473G congenic mouse strains differing in the single-nucleotide substitution C1473G in the Tph2 gene and activity of the encoded enzyme. Acute alcoholization led to a significant upregulation of the c-fos gene expression in the frontal cortex and striatum of B6-1473G mice and in the hippocampus of B6-1473C mice and caused a decrease in the index of serotonin metabolism in the nucleus accumbens in B6-1473C mice and in the hippocampus and striatum of B6-1473G mice, as well as to the decrease in the norepinephrine level in the hypothalamus of B6-1473C mice. Therefore, the C1473G polymorphism in the Tph2 gene has a significant effect of acute ethanol administration on the c-fos expression pattern and metabolism of biogenic amines in the mouse brain.
KEY WORDS: ethanol, C1473G polymorphism, tryptophan hydroxylase 2, c-fos gene, serotonin, dopamine, norepinephrine, mouseDOI: 10.1134/S000629792303001X
Abbreviations: 5-HT, serotonin; DA, dopamine; DOPAC, 3,4-dihydroxyphenylacetic acid; HVA, homovanillic acid; NE, norepinephrine; TPH2, tryptophan hydroxylase 2.
INTRODUCTION
The role of the serotonin (5-HT) brain system dysfunction in the pathophysiology of a broad spectrum of neuropsychological disorder, including alcohol-related ones, has been demonstrated in numerous studies [1, 2]. According to the published data, polymorphisms in genes encoding components of 5-HT brain system, such as 5-HT1B [3], 5-HT6 [4], and 5-HT2A [5] receptors, 5-HT transporter [6], and monoamine oxidase A [7], can cause behavioral disorders associated with alcohol consumption.
Tryptophan hydroxylase 2 (TPH2) is a key enzyme of serotonin synthesis in the brain. It was found that some polymorphisms in the human TPH2 gene are associated with the development of neuropsychological disorders, including depression and bipolar affective disorder [8, 9]. However, the role of this enzyme in the pathogenesis of alcohol-related diseases remains poorly studied. It was shown that alcoholic patients display increased levels of TPH2 mRNA and protein in the dorsal and median raphe nuclei [10]. The data of studies on the association between polymorphisms in the TPH2 gene and alcohol-related disorders are controversial [11-14]. Hence, development of appropriate animal models for studying the role of TPH2 gene in the effects of alcohol on the behavior and nervous system is of particular importance.
The single-nucleotide polymorphism (SNP) C1473G (rs33849125) in the mouse Tph2 gene is a functionally significant mutation resulting in the Pro447 to Arg447 substitution in the TPH2 protein, which leads to a twofold decrease in the enzyme activity and changes in the animal behavior [15, 16]. Earlier, we created congenic mouse strains B6-1473C and B6-1473G differing only by the presence of C1473G SNP in Tph2 [16]. It was demonstrated that the C1473G SNP is involved in the action of external stress factors (acute stress, short photoperiod) on the nervous system [17, 18]. We also found that acute alcohol administration affected differently the motor activity and anxiety behavior in B6-1473C and B6-1473G mice [19], although the mechanism of alcohol effect on the nervous system of these animals has not been studied. It is known that even a one-time injection of ethanol can change the metabolism of biogenic amines [20] and activate expression of early response genes (e.g., c-fos) in the brain [21] in a genotype-dependent manner.
The goal of our work was to investigate the effects of acute ethanol administration on the expression of early response gene c-fos and the content of 5-HT, norepinephrine (NE), dopamine (DA), and their metabolites in the brain structures of mice from congenic B6-1473C and B6-1473G strains differing in the presence of the C1473G SNP in the Tph2 gene.
MATERIALS AND METHODS
Animals. Adult male mice (10-12 weeks of age) of the congenic B6-1473C (C/C) and B6-1473G (G/G) strains were obtained by backcrossing male F1[CC57BR(G/G)xC57BL/6(C/C)] mice with C57BL/6 females for nine generations. The congenic B6-1473C and B6-1473G strains obtained on the C57BL/6 background differed only in the C1473G single-nucleotide substitution in the Tph2 gene [16]. The animals (body weight, 25 ± 0.7 g) were kept in plastic cages 40×30×15 cm under standard conditions (temperature, 20-22°C; relative humidity, 50-60%; 14 h light/10 h dark regime) with the ad libitum access to standard chow and water. The genotypes of animals used in the experiments were verified by genotyping as described in [16]. The studies were conducted at the Center for Genetic Resources of Laboratory Animals, Institute of Cytology and Genetics.
Ethanol administration and sacrificing the animals. Two days before the experiment, the animals were placed into individual cages. Ethanol (20%, 1.6 g per kg body weight) was injected intraperitoneally [22]; the mice of the control group were injected with physiological saline (0.9% NaCl). Forty minutes after ethanol injection, the animals were sacrificed by decapitation. The dose and the time of ethanol action were chosen based on the published data, as it was shown earlier that ethanol administration at a dose of 0.25-4 g per kg body weight results in the increase in the c-fos mRNA content within 1 h [21, 23]. On the other hand, this method of ethanol administration and its dose were sufficient to observed the effects of alcohol on the metabolism of biogenic amines [24].
Hypothalamus, frontal cortex, nucleus accumbens, hippocampus, striatum, substantia nigra, and midbrain were isolated on ice. These structures were selected because it was found that alcohol affects expression of early response genes in the hypothalamus, frontal cortex, hippocampus, and striatum [25, 26]. Raphe nuclei and substantia nigra are the major location of the cell bodies of serotoninergic [8] and dopaminergic [27] neurons, respectively. Nucleus accumbens and striatum receive mainly dopaminergic projections, while hypothalamus, frontal cortex, nucleus accumbens, hippocampus, and striatum receive serotoninergic projections [8, 27].
Brain structures were isolated based on information from the Mouse Brain Atlas [28]. The following coordinates were used for the fontal cortex: anterior-posterior (AP), from 1.6 to 2.8; lateral (L), from –2 to 2; section thickness, 1.5 mm. Hypothalamus was dissected based on the coordinates: AP, from 0.3 to –2.9; L, from –1 to 1; dorsal-ventral (DV), from 3.2 to 5.8. Both hippocampi were dissected from AP –0.8 to AP –2.9. The coordinates of the striatum were: AP, from 1.3 to –1.0; L, from –2.4 to –3.8 and from 2.4 to 3.8; DV, from 2.4 to 3.8. The coordinates of nucleus accumbens were: AP, from 1.8 to 1.2; L, from –1.7 to –0.5 and from 0.5 to 1.7; DV, from 4.2 to 3.6. To isolate the midbrain, we made the cranial cut in front of the superior colliculus (AP –3) and caudal cut in front of the rhomboid fossa (AP –7.3), after which the superior colliculus was removed. Substantia nigra was dissected according to the following coordinates: AP, from –2.7 to –3.4; L, from –1.2 to –2.0 and from 1.2 to 2.0; DV, from 3.6 to 4.4. The tissues were frozen in liquid nitrogen and stored at –70°C until being used in the experiment.
Two series of experiments were conducted. In the first series, brain tissues isolated from the animals injected with ethanol were used for RNA isolation (number of animals in the group, n = 6-7); in the second series, brain tissues were assayed for the content of biogenic amines by high-performance liquid chromatography (n = 7-8).
Total RNA isolation. Total RNA was isolated using TRIzol Reagent (Life Technologies, USA) and treated with RNase-free DNase (1000 U/ml; Promega, USA) as recommended by the manufacturer. RNA concentration was determined spectrophotometrically with a Nanodrop (USA). Isolated RNA was diluted with water to the concentration of 0.25 μg/μl and stored at –70°C. The presence of genomic DNA in the RNA preparations was assessed as described in [29, 30].
Reverse transcription (RT) was performed as described in [25]. Total RNA (8 μl or 1 μg) was mixed with 180 ng of random hexanucleotide primer (final primer concentration, 5 μM) and 2.25 μM sterile 1 M KCl in a total volume of 16 μl and denatured at 94°C for 5 min in a BIS M-120 cycler (BIS-N, Russia). Next, 15 μl of the reaction mixture containing 0.5 M Tris-HCl, pH 8.3 (0.63 μl), 4 mM dNTPs (3.63 μl), 0.1 M dithiothreitol (2.25 μl), 0.1 M MnCl2 (0.3 μl), 2000 U of MuMLV reverse transcriptase (Biosan, Russia), and 8 μl of diethyl pyrocarbonate-treated water was added and the resulting mixture (total volume, 31 μl) was incubated at 41°C for 60 min. Synthesized cDNA was stored at –20°C.
Quantitative polymerase chain reaction (qPCR). Gene expression was evaluated using a modification of the RT-qPCR method developed in our laboratory [24, 25]. The primers used for cDNA amplification (Table 1) were designed based on the sequences from the EMBL Nucleotide database using Oligoanalizer (https://eu.idtdna.com/calc/analyzer) and Ensemble (https://www.ensembl.org/index.html) software and synthesized by Biosan. The reaction mixture for qPCR contained 1 μl of cDNA and reagents from SYBR Green I Universal Reagent Kit (Sinto, Russia) as recommended by the manufacturer. The reaction was performed in a LightCycler 480 (Roche Applied Science, Switzerland) in the following regime: 95°C – 3 min; 40 cycles of 95°C – 10 s, annealing at the temperature shown in Table 1 – 30 s; 72°C – 30 s. Serial dilutions of genomic DNA with the concentrations of 0.125, 0.25, 0.5, 1, 2, 4, 8, 16, 32, and 64 ng/μl were amplified separately and used as external standards for plotting the calibration curve in the Ct (number of threshold cycle) – lg P (logarithm of standard DNA quantity) coordinates using software supplied with the LightCycler 480. To verify the specificity of amplification, the melting curves of qPCR products were obtained and analyzed for each reaction and each primer pair. Expression of the c-fos gene was presented as the ratio of the amount of c-fos cDNA to 100 copies of the DNA-dependent RNA polymerase 2 (rPol2) cDNA used as an internal standard [17, 29, 30].
Table 1. Primers used in the study

High-performance liquid chromatography (HPLC). The content of 5-HT and its metabolite 5-hydroxyindoleacetic acid (5-HIAA), DA and its metabolites 3,4-dihydroxyphenylacetic acid (DOPAC) and homovanillic acid (HVA), and NE in the brain was determined by HPLC as described in [31]. Brain tissue samples were homogenized in 150 μl of 0.6 M perchloric acid (HClO4) and centrifuged for 15 min at 4°C at 14,000 rpm to precipitate the protein. The supernatant was diluted with an equal volume of deionized water. The pellet was stored at –20°C for further protein concentration assay by the Bradford method [31]. The supernatant (20 μl) was analyzed using an HPLC system including an electrochemical detector (750 mV, DECADE IITM), glassy carbon VT03 flow cell (3 mm GC sb; Antec, Netherlands), CBM-20A controller, LC-20AD pump, SIL-20A autosampler, and DGU-20A5R degasser (Shimadzu, USA). Chromatography was carried out in the isocratic mode at a flow rate of 0.6 ml/min on a Luna C18 column (length, 75 mm; inner diameter, 4.6 mm; particle size, 5 μm) with a C8 precolumn (Phenomenex, USA). The eluent was 10% methanol (Chimmed, Russia) in 50 mM KH2PO4 (Sigma-Aldrich, USA) containing 1.4 mM 1-octanesulfonic acid sodium salt (Chimmed) and 0.05 mM EDTA (Sigma-Aldrich), pH 3.9. The calibration curves were plotted using serial dilutions containing 0.5, 1, and 2 ng of the analyzed compounds. The peaks in the chromatograms were quantified with the LabSolution LG/GC program (Shimadzu); the content of the assayed biogenic amines was determined from the calibration curves and expressed in ng/mg protein.
Statistical analysis. The results were presented as mean ± standard error of mean (SEM) and compared using the two-factor dispersion analysis ANOVA (factors “genotype” and “ethanol”), followed by the group comparison with the Fisher least significant difference (LSD) method using the STATISTICA 8.0 software. The normality of the samples was determined using the Kolmogorov–Smirnov test with the Lilliefors correction and the Shapiro–Wilk test. The differences were considered significant at p < 0.05.
RESULTS
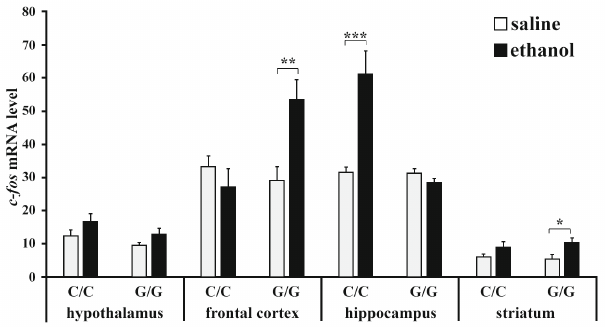
Expression of c-fos gene. ANOVA revealed that ethanol (F1,22 = 5.23; p < 0.05) and interaction between the “genotype” and “ethanol” factors (F1,22 = 9.85; p < 0.01) affected the content of c-fos mRNA in the frontal cortex, while the effect of genotype was at the trend level (F1,22 = 3.36, p = 0.08). Post hoc analysis showed that ethanol induced a significant increase in the c-fos expression in the frontal cortex in B6-1473G mice only (p < 0.01) (Fig. 1). Genotype (F1,22 = 24.15; p < 0.001), ethanol administration (F1,22 = 16.22; p < 0.001), and interaction of these factors (F1,22 = 23.74; p < 0.001) affected the c-fos mRNA level in the hippocampus. Group comparison revealed that ethanol caused upregulation of the c-fos expression in the hippocampus of B6-1473C mice only (p < 0.001) (Fig. 1). Ethanol (F1,22 = 8.52; p < 0.01), but not genotype (F1,22 < 1) or interaction of these two factors (F1,22 < 1) affected the content of c-fos mRNA in the striatum. The level of c-fos mRNA in the stratum of B6-1473G mice injected with ethanol was higher than in animals of the control group (p < 0.05). In the hypothalamus, c-fos expression was affected by ethanol (F1,20 = 4.59; p < 0.05) and genotype (at the trend level; F1,20 = 3.36; p = 0.08), but not by the interaction of these factors (F1,22 < 1), while group comparison revealed no differences between animals of different groups (Fig. 1).
Fig. 1. Effect of acute ethanol administration on the c-fos expression in the brain structures of B6-1473C (C/C) and B6-1473G (G/G) mice. Gene expression is shown as the ratio of c-fos cDNA to 100 copies of rPol2 cDNA (n = 6-7 in each group); * p < 0.05, ** p < 0.01, *** p < 0.001 compared to the control animal group.
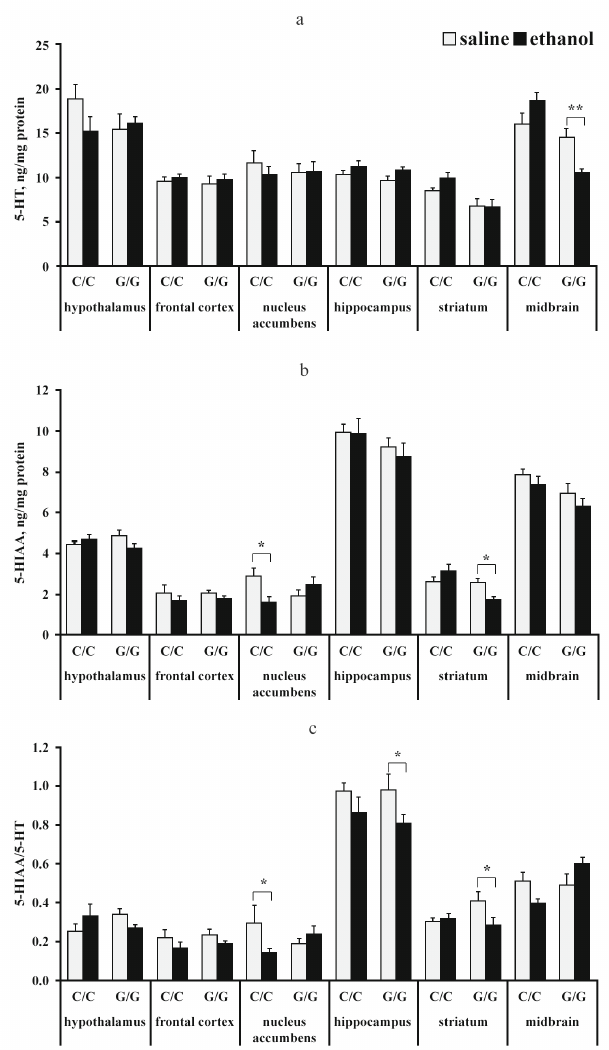
The content of serotonin and its metabolite 5-HIAA. The results of dispersion analysis of the effects of ethanol administration and genotype, as well as interaction of these factors, on the content of 5-HT, its metabolite 5-HIAA, and serotonin metabolism index (5-HIAA/5-HT) in the brain of B6-1473C and B6-1473G mice shown in Table 2. Group comparison revealed that after ethanol injection, the content of 5-HT in the midbrain decreased in B6-1473G mice (p < 0.01) and displayed a trend toward increase (p = 0.069) in B6-1473C mice (Fig. 2a). Post hoc analysis demonstrated a decrease in the 5-HIAA content in the nucleus accumbens of B6-1473C mice (p < 0.05) and striatum of B6-1473G mice (p < 0.05) injected with ethanol vs. control animals (Fig. 2b). Ethanol injection decreased serotonin metabolism index (5-HIAA/5-HT) in the nucleus accumbens of B6-1473C mice (p < 0.05) and in the hippocampus (p < 0.05) and striatum of B6-1473G mice (p < 0.05) (Fig. 2c). In the midbrain, this parameter showed the trend toward decrease in B6-1473C mice (p = 0.054) and toward increase in B6-1473G mice (p = 0.056) (Fig. 2c).
Fig. 2. Effect of acute ethanol administration on the content of 5-HT (a), its metabolite 5-HIAA (b), and serotonin metabolism index 5-HIAA/5-HT (c) in the brain structures of B6-1473C (C/C) and B6-1473G (G/G) mice (n = 7-8 in each group); * p < 0.05, ** p < 0.01 compared to the control group.
Table 2. ANOVA analysis of the effect of
“genotype” and “ethanol” factors and their
interaction on the 5-HT and 5-HIAA content and serotonin metabolism
index (5-HIAA/5-HT) in the brain structures of B6-1473C (C/C) and
B6-1473G (G/G) mice injected with ethanol
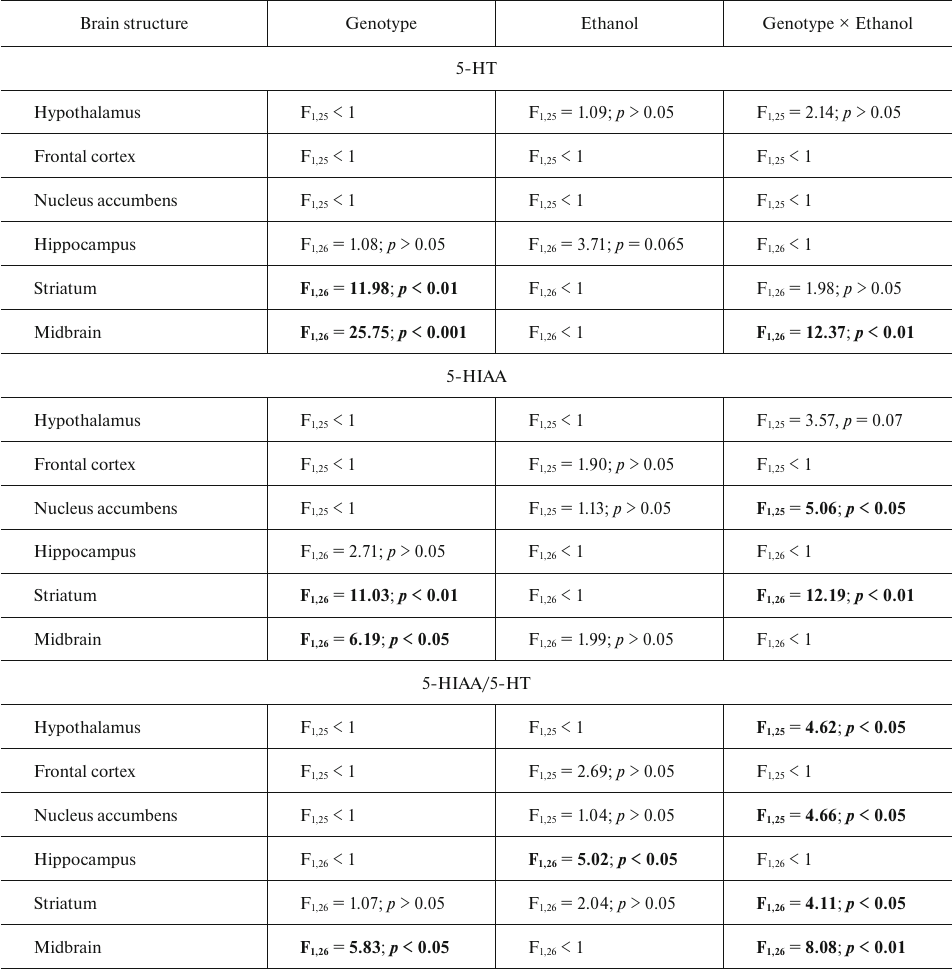
Note. Statistically significant results are shown in bold.
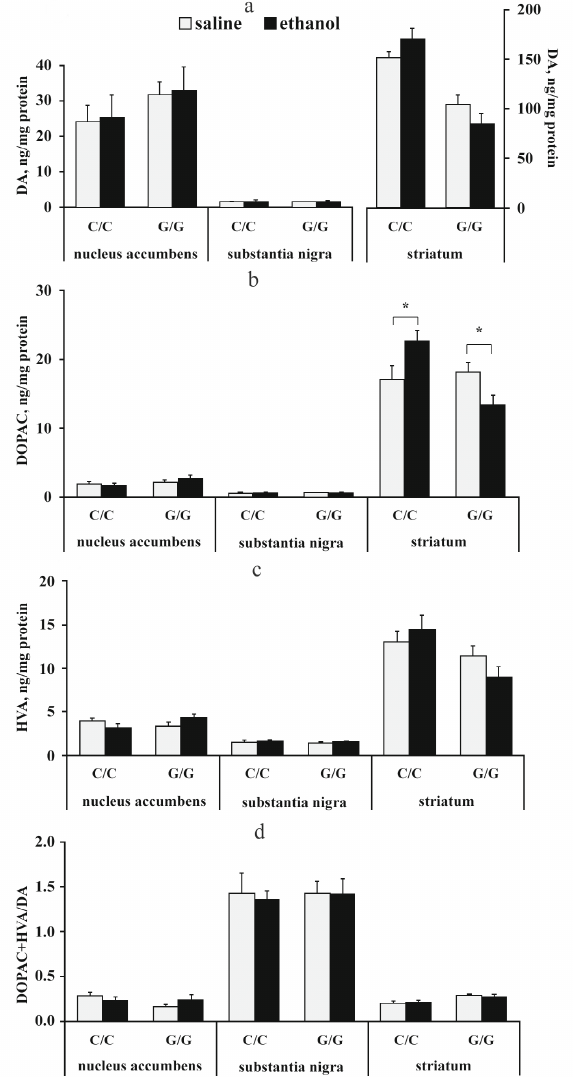
The content of dopamine (DA) and its metabolites DOPAC and HVA. The results of dispersion analysis of the effects of ethanol administration and genotype, as well as interaction of these factors, on the content of DA, its metabolites DOPAC and HVA, and dopamine metabolism index DOPAC + HVA/DA in the brain of B6-1473C and B6-1473G mice injected with ethanol are shown in Table 3. Comparison of groups revealed no differences in the DA content in all analyzed brain structures of the two mouse strains (p > 0.05) (Fig. 3a). However, post hoc analysis demonstrated that alcohol injection increased the DOPAC level in the striatum of B6-1473C mice (p < 0.05) and decreased it in B6-1473G mice (p < 0.05) (Fig. 3b). No differences in the HVA content and DA metabolism index were found in the mice of two strains (p > 0.05) (Fig. 3, c and d).
Fig. 3. Effect of acute ethanol administration on the content of DA (a), its metabolites DOPAC (b) and HVA (c), and DA metabolism index DOPAC + HVA/DA (d) in the brain structures of B6-1473C (C/C) and B6-1473G (G/G) mice (n = 7-8 in each group); * p < 0.05 compared to animals of the control group.
Table 3. ANOVA analysis of the effect of
“genotype” and “ethanol” factors and their
interaction on the content of DA, DOPAC, and HVA and dopamine
metabolism index DOPAC+HVA/DA in the brain structures of B6-1473C (C/C)
and B6-1473G (G/G) mice injected with ethanol
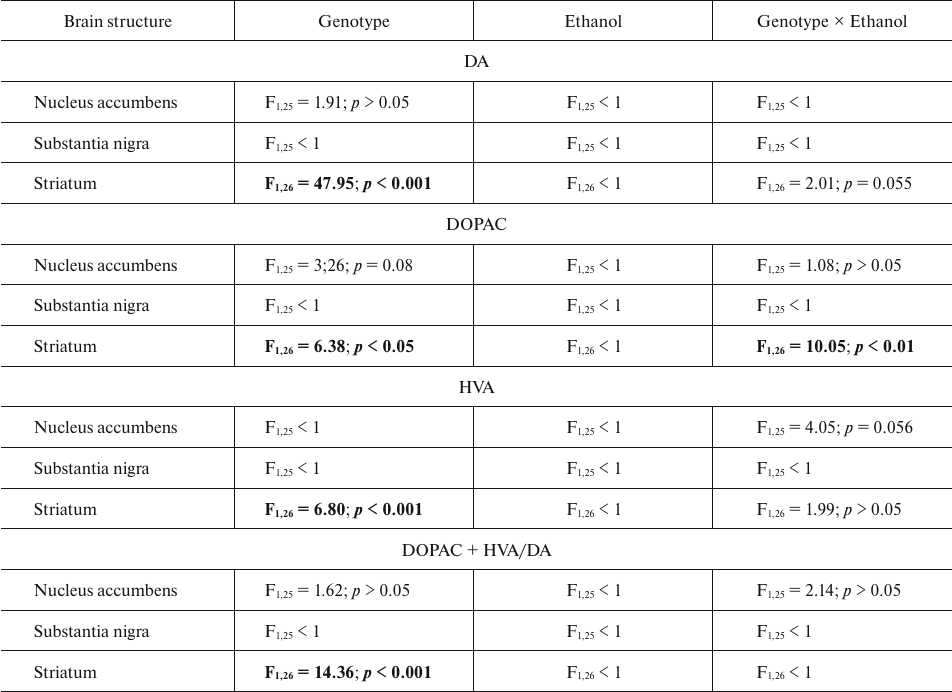
Note. Statistically significant results are shown in bold.
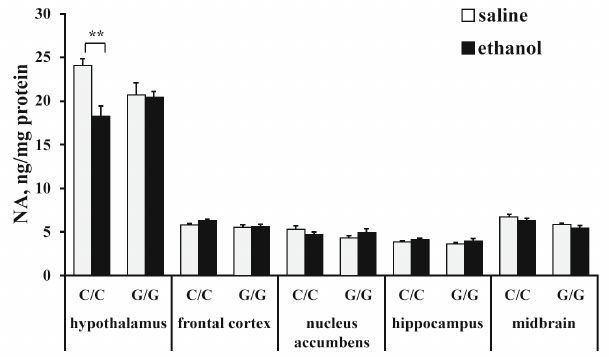
The content of norepinephrine (NE). ANOVA revealed that ethanol (F1,25 = 7,53; p < 0,05) and interaction between the “genotype” and “ethanol” factors (F1,25 = 6.64; p < 0.05), but not genotype (F1,25 < 1), affected the NE level in the hippocampus. Post hoc analysis demonstrated that ethanol injection induced a statistically significant decrease in the NE content in the hypothalamus in B6-1473C mice only (p < 0.01) (Fig. 4). Neither genotype (F1,26 = 3.93; p = 0.058), nor ethanol (F1,26 = 1.98; p > 0.05) or interaction between these factors (F1,26 < 1) affected NE content in the frontal cortex. Also, ANOVA revealed no effects of the genotype (F1,25 < 1), ethanol administration (F1,25 < 1), or interaction of these factors (F1,25 = 2.77; p > 0.05) on the NE content in the nucleus accumbens. Ethanol (F1,26 = 6.52; p < 0.05), but nor genotype (F1,26 = 1.88; p > 0.05) or interaction of the “ethanol” and “genotype” factors (F1,26 < 1) influenced the level of NE in the hippocampus, although in the hippocampus of B6-1473G mice, there was a trend toward the increase in the NE content (p = 0.66). ANOVA also showed the effect of the genotype (F1,25 = 15.61; p < 0.001), but not ethanol (F1,26 = 3,95; p = 0,058) or interaction between these factors (F1,26 < 1) in the midbrain. Group comparison revealed no differences in the NE content in the midbrain between the control and experimental mice (Fig. 4).
Fig. 4. Effect of acute ethanol administration on the NE content in the brain structures of B6-1473C (C/C) and B6-1473G (G/G) mice (n = 7-8 in each group); ** p < 0.01 compared to animals of the control group.
DISCUSSION
This study was aimed to reveal the role of the C1473G SNP in the Tph2 gene in the acute effects of ethanol on the mouse nervous system, including expression of the early response c-fos gene and metabolism of biogenic amines in the brain structures of mice from the congenic B6-1473C and B6-1473G strains. The mouse strains differed in the presence of the C1473G SNP in the Tph2 gene and, therefore, exhibited different activity of the encoded enzyme [16].
According to the published data, upregulation of the c-fos expression is an indicator of neuronal activity in the brain induced by various stress factors including acute alcohol intoxication [25, 32, 33]. It was observed that the level of the c-fos mRNA increases within 1 h after ethanol administration at a dose of 0.25-4 g/kg body weight [21, 23]. However, some researchers argue that even if the upregulation of the c-fos expression and synthesis of its protein product may provide an important information on the activation of cellular signaling, the absence of expression upregulation does not necessarily indicate the lack of neuronal activation after experimental manipulations [34]. We found that changes in the c-fos expression pattern induced by alcohol administration differed in different brain structures. Thus, alcohol increased the levels of c-fos mRNA in the hippocampus of B6-1473C mice (with normal TPH2 activity) and in the frontal cortex and striatum of B6-1473G mice (with the TPH2 activity decreased by 50%). Interestingly, in the earlier experiments on the effect of acute emotional stress (30 min), expression of c-fos significantly increased in the hippocampus and cortex of mice from both strains [17].
Multiple studies have shown that acute alcohol administration might either increase or decrease the content of 5-HT and its main metabolite 5-HIAA in the entire brain and/or its regions, or might have no effect on the level of these compounds in the brain [35, 36]. Such discrepancies can be explained by the fact that the authors of such studies have used different animals, animal strains, and experimental designs (ethanol dose, method of administration, time intervals, etc.). In our study, we observed significant alcohol-induced changes in the serotonin metabolism in B6-1473G mice, such as decrease in the 5-HT content in the raphe nuclei in the midbrain and in the content of 5-HIAA in the striatum, as well as in the serotonin metabolism index in the hippocampus and striatum. In B6-1473C mice, the most susceptible brain structure was nucleus accumbens, where acute alcohol administration caused a decrease in the 5-HIAA content and serotonin metabolism index. Notably, the serotonin metabolism index in the raphe nuclei changed in the opposite directions in the two mouse strains, i.e., decreased in B6-1473C mice and increased in B6-1473G mice (although at the trend level). Since raphe nuclei are the major area containing cell bodies of serotoninergic neurons [8], we explain the observed effect of ethanol by the features of the serotonin system in the brain of B6-1473G mice. It should be noted that despite the fact that the substitution of cytosine with guanine at position 1473 in the Tph2 gene results in a significant decrease in the TPH2 activity, we found no differences in the 5-HT content between the control groups of B6-1473G and B6-1473C mice, which is in agreement with the results of earlier studies [16, 37]. This can be explained by the development of compensatory processes in the serotonin system in the brain of B6-1473G mice (due to the decrease in the TPH2 activity). One of these processes is desensitization of presynaptic 5-HT1A receptors located in the raphe nuclei and regulating serotonin release by the feedback mechanism [38]. The mechanisms underlying the effect of acute alcohol administration of the 5-HT release remain unclear, although it is believed that they involve the activity of presynaptic 5-HT1A autoreceptors [39]. It was suggested that acute alcohol administration in B6-1473G mice disrupts the compensatory processes in the midbrain, making serotonin brain system more susceptible to alcohol. This hypothesis is corroborated by the data on the alcohol-induced increase in the sensitivity of 5-HT1A autoreceptors [40]. It should be noted that the 30-min restriction stress produced a significant effect on the 5-HT metabolism in B6-1473C mice [17]. Taken together with the results of our work, these data suggest an existence of different mechanisms regulating the effects of acute emotional stress on the brain serotonin system and expression of the early response c-fos in B6-1473G and B6-1473C mice.
Other important neurotransmitters involved in the control of ethanol-induced effects on the nervous system are DA and NE [41, 42]. We found that alcohol administration caused the opposite effects on the content of DA metabolite DOPAC in the striatum, i.e., decreased it in B6-1473G mice and increased in B6-1473C mice. The latter result can indirectly indicate induction by ethanol of the increase in the content of extracellular DA in the striatum, which is in agreement with the published data [43]. It was shown earlier that the C1473G SNP in the Tph2 gene affects the metabolism of the DA brain system resulting in the decrease in the DA and DOPAC levels in the midbrain and striatum [44], although the mechanism of this effect also remains unknown. It is possible that alterations in the dopamine system in B6-1473G mice determine the differences in the response of this brain system to alcohol in the two strains of mice. Also, we observed an alcohol-induced decrease in the NE content in the hypothalamus in B6-1473C mice only, which correlated with the data on a higher sensitivity of these animals to acute stress [17].
Therefore, the C1473G SNP in the Tph2 gene is involved in the regulation of the effects of acute alcohol administration on the expression of the early response c-fos gene and metabolism of important neurotransmitters, such as serotonin and catecholamines. Our results suggest that B6-1473G and B6-1473C mice can be used in studying the regulation of alcohol action on the central nervous system.
Contributions. D.V.B. and A.V.K. developed the concept, supervised the study, and discussed the results; D.V.B., D.V.F., and V.S.N. carried out the experiments; D.V.B. wrote the manuscript.
Funding. This work was supported by the Russian Science Foundation (project no. 21-15-00051). The animals’ maintenance was funded by a Budget Project FWNR-2022-0023.
Ethics declarations. The authors declare no conflict of interest. All procedures were performed in compliance with the National Institute of Health Guide for the Care and Use of Laboratory Animals (NIH Publications no. 80023, 1996) and the order no. 119 from 01.04.2016 “On the establishment of rules for the proper laboratory practice” (no. 43232, registered on 15.08.2016) by the Ministry of Health of the Russian Federation. The experiments and animal maintenance procedures were approved by the Bioethics Committee of the Institute of Cytology and Genetics.
REFERENCES
1.Sari, Y., Johnson, V. R., and Weedman, J. M. (2011)
Role of the serotonergic system in alcohol dependence: from animal
models to clinics, Prog. Mol. Biol. Transl. Sci., 98,
401-443, doi: 10.1016/B978-0-12-385506-0.00010-7.
2.Marcinkiewcz, C. A. (2015) Serotonergic systems in
the pathophysiology of ethanol dependence: relevance to clinical
alcoholism, ACS Chem. Neurosci., 6, 1026-1039, doi:
10.1021/cn5003573.
3.Lappalainen, J., Long, J. C., Eggert, M., Ozaki,
N., Robin, R. W., Brown, G. L., Naukkarinen, H., Virkkunen, M.,
Linnoila, M., and Goldman, D. (1998) Linkage of antisocial alcoholism
to the serotonin 5-HT1B receptor gene in 2 populations, Arch. Gen.
Psychiatry, 55, 989-994, doi:
10.1001/archpsyc.55.11.989.
4.Bonasera, S. J., Chu, H. M., Brennan, T. J., and
Tecott, L. H. (2006) A null mutation of the serotonin 6 receptor alters
acute responses to ethanol, Neuropsychopharmacology, 31,
1801-1813, doi: 10.1038/sj.npp.1301030.
5.Wrzosek, M., Jakubczyk, A., Wrzosek, M., Matsumoto,
H., Łukaszkiewicz, J., Brower, K. J., and Wojnar, M. (2012)
Serotonin 2A receptor gene (HTR2A) polymorphism in alcohol-dependent
patients, Pharmacol. Rep., 64, 449-453, doi:
10.1016/s1734-1140(12)70787-9.
6.Boyce-Rustay, J. M., Wiedholz, L. M., Millstein, R.
A., Carroll, J., Murphy, D. L., Daws, L. C., and Holmes, A. (2006)
Ethanol-related behaviors in serotonin transporter knockout mice,
Alcohol. Clin. Exp. Res., 30, 1957-1965, doi:
10.1111/j.1530-0277.2006.00241.x.
7.Popova, N. K., Vishnivetskaya, G. B., Ivanova, E.
A., Skrinskaya, J. A., and Seif, I. (2000) Altered behavior and alcohol
tolerance in transgenic mice lacking MAO A: a comparison with effects
of MAO A inhibitor clorgyline, Pharmacol. Biochem. Behav.,
67, 719-727, doi: 10.1016/s0091-3057(00)00417-2.
8.Kulikova, E. A., and Kulikov, A. V. (2019)
Tryptophan hydroxylase 2 as a therapeutic target for psychiatric
disorders: focus on animal models, Expert. Opin. Ther. Targets,
23, 655-667, doi: 10.1080/14728222.2019.1634691.
9.Zhang, X., Gainetdinov, R. R., Beaulieu, J. M.,
Sotnikova, T. D., Burch, L. H., Williams, R. B., Schwartz, D. A.,
Krishnan, K. R., and Caron, M. G. (2005) Loss-of-function mutation in
tryptophan hydroxylase-2 identified in unipolar major depression,
Neuron, 45, 11-16, doi: 10.1016/j.neuron.2004.12.014.
10.Bach, H., Arango, V., Kassir, S. A., Tsaava, T.,
Dwork, A. J., Mann, J. J., and Underwood, M. D. (2014) Alcoholics have
more tryptophan hydroxylase 2 mRNA and protein in the dorsal and median
raphe nuclei, Alcohol. Clin. Exp. Res., 38, 1894-1901,
doi: 10.1111/acer.12414.
11.Zupanc, T., Pregelj, P., Tomori, M., Komel, R.,
and Paska, A. V. (2011) TPH2 polymorphisms and alcohol-related suicide,
Neurosci. Lett., 490, 78-81, doi:
10.1016/j.neulet.2010.12.030.
12.Bragatti, J. A., Bandeira, I. C., de Carvalho, A.
M., Abujamra, A. L., Leistner-Segal, S., and Bianchin, M. M. (2014)
Tryptophan hydroxylase 2 (TPH2) gene polymorphisms and psychiatric
comorbidities in temporal lobe epilepsy, Epilepsy Behav.,
32, 59-63, doi: 10.1016/j.yebeh.2014.01.007.
13.Zill, P., Preuss, U. W., Koller, G., Bondy, B.,
and Soyka, M. (2007) SNP- and haplotype analysis of the tryptophan
hydroxylase 2 gene in alcohol-dependent patients and alcohol-related
suicide, Neuropsychopharmacology, 32, 1687-1694, doi:
10.1038/sj.npp.1301318.
14.Gacek, P., Conner, T. S., Tennen, H., Kranzler,
H. R., and Covault, J. (2008) Tryptophan hydroxylase 2 gene and alcohol
use among college students, Addict. Biol., 13, 440-448,
doi: 10.1111/j.1369-1600.2008.00118.x.
15.Zhang, X., Beaulieu, J. M., Sotnikova, T. D.,
Gainetdinov, R. R., and Caron, M. G. (2004) Tryptophan hydroxylase-2
controls brain serotonin synthesis, Science, 305, 217,
doi: 10.1126/science.1097540.
16.Osipova, D. V., Kulikov, A. V., and Popova, N. K.
(2009) C1473G polymorphism in mouse tph2 gene is linked to tryptophan
hydroxylase-2 activity in the brain, intermale aggression, and
depressive-like behavior in the forced swim test, J. Neurosci.
Res., 87, 1168-1174, doi: 10.1002/jnr.21928.
17.Bazhenova, E. Y., Bazovkina, D. V., Kulikova, E.
A., Fursenko, D. V., Khotskin, N. V., Lichman, D. V., and Kulikov, A.
V. (2017) C1473G polymorphism in mouse tryptophan hydroxylase-2 gene in
the regulation of the reaction to emotional stress, Neurosci.
Lett., 640, 105-110, doi: 10.1016/j.neulet.2017.01.010.
18.Bazhenova, E. Y., Fursenko, D. V., Kulikova, E.
A., Khotskin, N. V., Sinyakova, N. A., and Kulikov, A. V. (2019) Effect
of photoperiodic alterations on depression-like behavior and the brain
serotonin system in mice genetically different in tryptophan
hydroxylase 2 activity, Neurosci. Lett., 699, 91-96, doi:
10.1016/j.neulet.2019.01.041.
19.Bazovkina, D. V., Lichman, D. V., and Kulikov, A.
V. (2015) The C1473G polymorphism in the Tryptophan hydroxylase-2 gene:
involvement in ethanol-related behavior in mice, Neurosci.
Lett., 589, 79-82, doi: 10.1016/j.neulet.2015.01.043.
20.Boone, E. M., Cook, M. N., Hou, X., and Jones, B.
C. (1997) Sex and strain influence the effect of ethanol on central
monoamines, J. Stud. Alcohol, 58, 590-599, doi:
10.15288/jsa.1997.58.590.
21.Hitzemann, B., and Hitzemann, R. (1997) Genetics
ethanol and the Fos response: a comparison of the C57BL/6J and DBA/2J
inbred mouse strains, Alcohol. Clin. Exp. Res., 21,
1497-1507.
22.Raymond, J. S., Wilson, B. B., Tan, O.,
Gururajan, A., and Bowen, M. T. (2019) Acute alcohol exposure
dose-dependently alleviates social avoidance in adolescent mice and
inhibits social investigation in adult mice, Psychopharmacology,
236, 3625-3639, doi: 10.1007/s00213-019-05335-8.
23.Ogilvie, K. M., Lee, S., and Rivier, C. (1998)
Divergence in the expression of molecular markers of neuronal
activation in the parvocellular paraventricular nucleus of the
hypothalamus evoked by alcohol administration via different routes,
J. Neurosci., 18, 4344-4352.
24.Nevo, I., and Hamon, M. (1995) Neurotransmitter
and neuromodulatory mechanisms involved in alcohol abuse and
alcoholism, Neurochem. Int., 26, 305-342, doi:
10.1016/0197-0186(94)00139-l.
25.Vilpoux, C., Warnault, V., Pierrefiche, O.,
Daoust, M., and Naassila, M. (2009) Ethanol-sensitive brain regions in
rat and mouse: a cartographic review, using immediate early gene
expression, Alcohol. Clin. Exp. Res., 33, 945-969, doi:
10.1111/j.1530-0277.2009.00916.x.
26.Abrahao, K. P., Salinas, A. G., and Lovinger, D.
M. (2017) Alcohol and the brain: neuronal molecular targets, synapses,
and circuits, Neuron, 96, 1223-1238, doi:
10.1016/j.neuron.2017.10.032.
27.Ford, C. P. (2014) The role of D2-autoreceptors
in regulating dopamine neuron activity and transmission,
Neuroscience, 282, 13-22, doi:
10.1016/j.neuroscience.2014.01.025.
28.Slotnick, B. M., and Leonard, C. M. (1975) A
Stereotaxic Atlas of the Albino Mouse Forebrain, U. S. Dept. of
Health, Education and Welfare, Rockville, 174 p.
29.Naumenko, V. S., and Kulikov, A. V. (2006)
Quantitative assay of 5-HT(1A) serotonin receptor gene expression in
the brain, Mol. Biol. (Mosk), 40, 37-44, doi:
10.1134/s0026893306010079.
30.Naumenko, V. S., Osipova, D. V., Kostina, E. V.,
and Kulikov, A. V. (2008) Utilization of a two-standard system in
real-time PCR for quantification of gene expression in the brain, J.
Neurosci. Methods, 170, 197-203, doi:
10.1016/j.jneumeth.2008.01.008.
31.Bazovkina, D., Naumenko, V., Bazhenova, E., and
Kondaurova, E. (2021) Effect of central administration of brain-derived
neurotrophic factor (BDNF) on behavior and brain monoamine metabolism
in new recombinant mouse lines differing by 5-HT1A receptor
functionality, Int. J. Mol. Sci., 22, 11987, doi:
10.3390/ijms222111987.
32.Robinson, S. L., Dornellas, A., Burnham, N. W.,
Houck, C. A., Luhn, K. L., Bendrath, S. C., Companion, M. A., Brewton,
H. W., Thomas, R. D., Navarro, M., and Thiele, T. E. (2020) Distinct
and overlapping patterns of acute ethanol-induced c-Fos activation in
two inbred replicate lines of mice selected for drinking to high blood
ethanol concentrations, Brain Sci., 10, 988, doi:
10.3390/brainsci10120988.
33.Smith, M. L., Li, J., Cote, D. M., and Ryabinin,
A. E. (2016) Effects of isoflurane and ethanol administration on c-Fos
immunoreactivity in mice, Neuroscience, 316, 337-343,
doi: 10.1016/j.neuroscience.2015.12.047.
34.McBride, W. J. (2002) Central nucleus of the
amygdala and the effects of alcohol and alcohol-drinking behavior in
rodents, Pharmacol. Biochem. Behav., 71, 509-515, doi:
10.1016/s0091-3057(01)00680-3.
35.LeMarquand, D., Pihl, R. O., and Benkelfat, C.
(1994) Serotonin and alcohol intake, abuse, and dependence: findings of
animal studies, Biol. Psychiatry, 36, 395-421, doi:
10.1016/0006-3223(94)91215-7.
36.Badawy, A. A. (2002) Tryptophan metabolism in
alcoholism, Nutr. Res. Rev., 15, 123-152, doi:
10.1079/NRR200133.
37.Siesser, W. B., Zhang, X., Jacobsen, J. P.,
Sotnikova, T. D., Gainetdinov, R. R., and Caron, M. G. (2010)
Tryptophan hydroxylase 2 genotype determines brain serotonin synthesis
but not tissue content in C57Bl/6 and BALB/c congenic mice,
Neurosci. Lett., 481, 6-11, doi:
10.1016/j.neulet.2010.06.035.
38.Berger, S. M., Weber, T., Perreau-Lenz, S., Vogt,
M. A., Gartside, S. E., Maser-Gluth, C., Lanfumey, L., Gass, P.,
Spanagel, R., and Bartsch, D. (2012) A functional Tph2 C1473G
polymorphism causes an anxiety phenotype via compensatory changes in
the serotonergic system, Neuropsychopharmacology, 37,
1986-1998, doi: 10.1038/npp.2012.46.
39.Kirby, L. G., Zeeb, F. D., and Winstanley, C. A.
(2011) Contributions of serotonin in addiction vulnerability,
Neuropharmacology, 61, 421-432, doi:
10.1016/j.neuropharm.2011.03.022.
40.Kelaï, S., Renoir, T., Chouchana, L.,
Saurini, F., Hanoun, N., Hamon, M., and Lanfumey, L. (2008) Chronic
voluntary ethanol intake hypersensitizes 5-HT1A
autoreceptors in C57BL/6J mice, J. Neurochem., 107,
1660-1670, doi: 10.1111/j.1471-4159.2008.05733.x.
41.Melis, M., Diana, M., Enrico, P., Marinelli, M.,
and Brodie, M. S. (2009) Ethanol and acetaldehyde action on central
dopamine systems: mechanisms, modulation, and relationship to stress,
Alcohol, 43, 531-539, doi:
10.1016/j.alcohol.2009.05.004.
42.Weinshenker, D., and Schroeder, J. P. (2007)
There and back again: a tale of norepinephrine and drug addiction,
Neuropsychopharmacology, 32, 1433-1451, doi:
10.1038/sj.npp.1301263.
43.Tang, A., George, M. A., Randall, J. A., and
Gonzales, R. A. (2003) Ethanol increases extracellular dopamine
concentration in the ventral striatum in C57BL/6 mice, Alcohol.
Clin. Exp. Res., 27, 1083-1089, doi:
10.1097/01.ALC.0000075825.14331.65.
44.Sinyakova, N. A., Bazhenova, E. Y., Kulikova, E.
A., Fursenko, D. V., and Kulikov, A. V. (2020) Effect of the C1473G
polymorphic variant of the tryptophan hydroxylase 2 gene and
photoperiod length on the dopamine system of the mouse brain [in
Russian], Mol. Biol. (Mosk), 54, 60-68, doi:
10.31857/S0026898420010140.