REVIEW: Sphingolipid Metabolism in Tumor Cells
Vadim S. Pokrovsky1,a*, Veronika I. Ivanova-Radkevich1, and Olga M. Kuznetsova1
1People’s Friendship University of Russia (RUDN University), 117198 Moscow, Russia* To whom correspondence should be addressed.
Received February 25, 2023; Revised May 16, 2023; Accepted May 17, 2023
Sphingolipids are a diverse family of complex lipids typically composed of a sphingoid base bound to a fatty acid via amide bond. The metabolism of sphingolipids has long remained out of focus of biochemical studies. Recently, it has been attracting an increasing interest of researchers because of different and often multidirectional effects demonstrated by sphingolipids with a similar chemical structure. Sphingosine, ceramides (N-acylsphingosines), and their phosphorylated derivatives (sphingosine-1-phosphate and ceramide-1-phosphates) act as signaling molecules. Ceramides induce apoptosis and regulate stability of cell membranes and cell response to stress. Ceramides and sphingoid bases slow down anabolic and accelerate catabolic reactions, thus suppressing cell proliferation. On the contrary, their phosphorylated derivatives (ceramide-1-phosphate and sphingosine-1-phosphate) stimulate cell proliferation. Involvement of sphingolipids in the regulation of apoptosis and cell proliferation makes them critically important in tumor progression. Sphingolipid metabolism enzymes and sphingolipid receptors can be potential targets for antitumor therapy. This review describes the main pathways of sphingolipid metabolism in human cells, with special emphasis on the properties of this metabolism in tumor cells.
KEY WORDS: sphingolipids, tumor, ceramide, dihydroceramide, sphingomyelin, sphingosine-1-phosphate, proliferation, cancerDOI: 10.1134/S0006297923070015
Abbreviations: ARS, arylsulfatase; Cer, ceramide; CERase, ceramidase; CERK, ceramide kinase; CERS, ceramide synthase; CST, cerebroside sulfotransferase; CERT, ceramide transporter; C1P, ceramide-1-phosphate; DEGS, dihydroceramide desaturase; dhCer, dihydroceramide; dhS1P, dihydrosphingosine-1-phosphate; ER, endoplasmic reticulum; GalCer, galactosylceramide; GALC, galactosyl ceramidase; GalCERS, galactosylcerebroside synthase; GluCer, glucosylceramide; GluCERase, glucosylceramidase; GluCERS, glucosylcerebroside synthase; PA, phosphatidic acid; SGPP, sphingosine-1-phosphate phosphatase; SM, sphingomyelin; SMase, sphingomyelinase; SMS, sphingomyelin synthase; Sph, sphingosine; SPHK, sphingosine kinase; S1P, sphingosine-1-phosphate; S1PL, sphingosine-1-phosphate lyase, TNF-α, tumor necrosis factor α.
INTRODUCTION
For a long time, sphingolipid metabolism has been neglected in classical biochemistry and its investigation has remained secondary to the studies of lipids and lipidomics. Unlike triacylglycerols, sphingolipids are not essential energy sources and produce much less effect on the organism physiological state compared to lipids (e.g., cholesterol and steroid hormones), which kept them “in the shadow” of other lipids. However, understanding the signaling role and effect of sphingolipids on the assembly of cell membrane lipid rafts has been increasingly growing over recent decades. The studies of sphingolipids are primarily focused on their metabolism and function in neurons because impairments of sphingolipid metabolism often manifest as neurological symptoms.
Healthy cells maintain a balance between different sphingolipids, which ensures normal cell growth, division, differentiation, as well as death of unnecessary cells. This balance is created by enzymes involved in sphingolipid synthesis and degradation. Changes in the expression and activity of specific enzymes can result in the impaired sphingolipid balance, leading to the stimulation of cell proliferation, migration, invasion, and emergence of chemoresistance, or, conversely, to the activation of apoptosis.
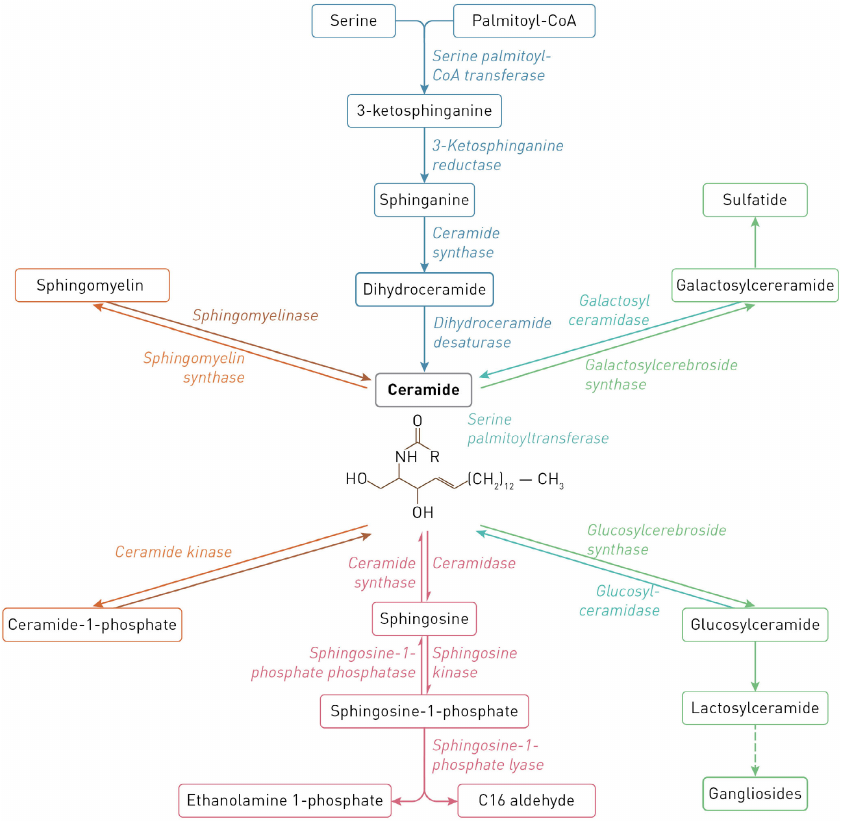
Reactions of sphingolipid biosynthesis and catabolism are commonly described as the sphingolipid (or ceramide, Cer) cycle. Most reactions of sphingolipid synthesis are reversible and take place in the endoplasmic reticulum (ER), on the ER membrane, and in the Golgi apparatus and mitochondria. Complex sphingolipids are catabolized in the lysosomes by lysosomal enzymes [1]. The scheme of metabolism of major cell sphingolipids in human cells and enzymes catalyzing reactions of sphingolipid synthesis and degradation are shown in Fig. 1.
Fig. 1. Scheme of sphingolipid metabolism.
A growing body of evidence on the role of sphingolipids in the metabolism and functioning of tumor cells has been reflected in the increasing number of publications on this topic that can be found in PubMed using the query “sphingolipids and cancer” (as of February 6, 2023, 8155 scientific publications were found). The first study appeared in 1958; the number of published reports peaked in 2020, 2021, and 2022 (366, 361, and 367 articles, respectively).
This expanding comprehension of the role of sphingolipid metabolism in tumor cells has motivated some researchers to systematize available data, which resulted in the publications of several reviews over the past three years. Most of these articles discuss sphingolipids and their signaling role as potential targets for the antitumor therapy in lung cancer [2], breast cancer [3], lymphomas [4], multiple myeloma [5], gliomas [6], and tumors of the gastrointestinal tract [7]).
Some reports assess the role of specific enzymes (ceramide kinase [8], β-galactosylceramidase [9]) in the sphingolipid metabolism in tumor cells or are dedicated to sphingolipid metabolism in a particular cell type, e.g., hepatocellular carcinoma cells [5]. Several well-illustrated reviews describe the overall sphingolipid metabolism [1, 10]; however, their focus is shifted mainly toward discussing individual elements of this metabolism as potential therapeutic targets.
The goal of our article was to systematize the data on sphingolipid metabolism with special emphasis of the features of this metabolism in tumor cells using a classical biochemical approach. We described the properties of enzymes involved in the synthesis and catabolism of major sphingolipids and their role in the mechanisms underlying survival and death of tumor cell. Because most classical biochemistry textbooks lack the section on sphingolipid metabolism, this review, along with its scientific message, might be used as a reference and educational source to provide young investigators with a deeper insight into this complex topic.
RELATIONSHIP BETWEEN THE ACTIVITY OF SPHINGOLIPID SYNTHESIS
ENZYMES AND TUMOR PROGRESSION
Ceramide synthesis. The three major pathways maintaining the concentration of ceramide (Cer) in mammalian cells are:
- de novo synthesis from serine and palmitoyl-SCoA by enzymes located on the cytoplasmic side of the ER membrane;
- catabolism of complex sphingolipids, e.g., sphingomyelin (SM), by sphingomyelinases (SMases) or phospholipase C that takes place on the plasma membrane, inside the Golgi apparatus, and presumably, in the mitochondria;
- catabolism of glucocerebrosides (GluCers) and galactocerebrosides (GalCers) in lysosomes.
De novo synthesis of Cers includes four sequential reactions (Fig. 1) [11]. Two enzymes of this metabolic pathway – ceramide synthase (CERS) and dihydroceramide desaturase – have been found to play an important role in the regulation of cell growth, proliferation, and development of antitumor response. CERS catalyzes the synthesis of dihydroceramide (dhCer) and is represented by six isoforms that add acyl residues differing in the length of the hydrocarbon chain and the number of double bonds (Table 1) [12].
Table 1. Human CerSase isoforms [12]
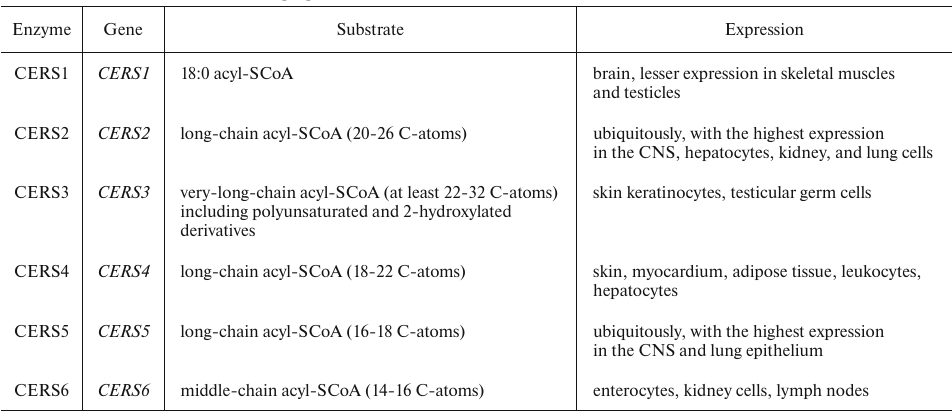
Because of the lower tissue concentrations compared to Cer, dhCers have been long believed to be inert lipids. Technological advances in lipidomics and availability of pharmacological inhibitors and transgenic models have led to the discovery of new properties and functions of dhCers, many of which were different or even opposite to the functions of Cers.
Changes in the dhCer content affect the lipid composition of organelle membranes, thus provoking a response to oxidative and ER stresses. In 2006, it was reported [13] that both short- and long-chain dhCers inhibit Cer-mediated pore formation in the outer mitochondrial membrane, a critical step in the apoptosis progression. As demonstrated in vitro and in vivo, dhCers accumulate in an HIF (hypoxia-inducible factor)-independent manner and suppress cell proliferation. The effect of hypoxia on dhCer synthesis was confirmed in vivo. Hypoxia decreased the content of Cers, but increased the amount of dhCers. dhCers have been characterized as molecules with antiproliferative and immunoregulatory properties. Some dhCer species produce a bidirectional effect: at high concentration, they elicit oxidative stress, while their synthesis is induced by the oxidative stress [14].
Cer is formed from dhCer by dihydroceramide desaturase (DEGS). In human, two DEGS isoforms have been identified – DEGS1 and DEGS2 (Table 2).
Table 2. Human DEGS isoforms [15-17]

The data obtained by Guo et al. [18] showed that DEGS2 plays an oncogenic role in colorectal cancer, whereas m6A-dependent changes in DEGS2 promote tumor progression and metastasis via downregulation of Cer synthesis. Moreover, DEGS2 expression is markedly increased in colon cancer vs. normal tissues and correlates with tumor response to immunotherapy and poor prognosis. Hence, DEGS2 can serve as a potential prognostic factor and therapeutic target in colon cancer.
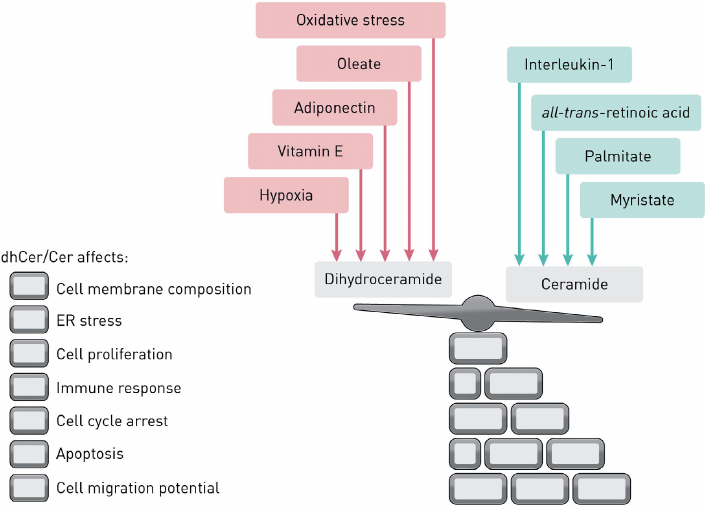
The activity of DEGS1 can be affected by the exposure to various modulators resulting in the altered dhCer/Cer ratio (Fig. 2), which impacts various cellular processes, such as apoptosis, cell proliferation, autophagy, and insulin resistance.
Fig. 2. Factors affecting the dhCer/Cer balance.
Hernández-Tiedra et al. [19] showed that the shift in the dhCer/Cer ratio toward dhCer activated autophagy and apoptosis in U87MG cells (human glioma cell line) and inhibited tumor growth. The authors suggested that exposure to delta-9-tetrahydrocannabinol can partially inhibit DEGS1.
Hydrophobic Cer molecules produced by DEGS assemble into Cer-rich microdomains that stabilize membrane lipid rafts, which activates signaling cascades by facilitating protein–protein interactions (including dimerization of receptor proteins) and lowering the activation threshold of signaling pathways. This effect of Cer can be viewed, at least partially, as a general mechanism for reshaping membrane-embedded signaling molecules [3, 20].
Excessive accumulation of Cers can be toxic. Normal cellular concentrations of free Cers are low, except in skin keratinocytes containing relatively high amounts of these compounds. Most cells rapidly utilize Cers in the synthesis of complex sphingolipids. As mentioned above, de novo Cer generation occurs on the cytoplasmic side of the ER membrane, whereas the synthesis of complex sphingolipids takes place in the Golgi apparatus [1, 14].
Cers are transported in the content of vesicles or by non-vesicular transport in an ATP-dependent or ATP-independent manner. Non-vesicular transport relies on transport proteins such as the cytosolic 68-kDa ceramide transporter (CERT) containing the Cer-binding site. This protein is strictly specific to Cer and does not interact with other lipids (cholesterol, phospholipids, etc.), although it exhibits a weak affinity toward diacylglycerols. The efficiency of transfer depends on the length of acyl radicals. The most efficiently transported Cers are Cer(C14:0-C20:0), whereas the transport rate for Cer(C22:0) and Cer(C24:1) is ∼40% of that for shorter Cer species. There is almost no transport of Cer(C24:0). It is believed that the preferential recognition of Cer acyl radicals by CERT reflects the chemical composition of radicals in SM and GluCer [21]. Once in the cytosolic leaflet of the Golgi apparatus membrane, Cer molecule is transferred to the inner layer, where it is used as a substrate in the synthesis of complex sphingolipids. This process relies mostly on Cers(C16-18) that are selectively extracted by CERT from cell membranes and redirected to the Golgi apparatus. Cers with longer fatty acids [Cer(C20-26)] are retained in the membranes for longer periods of time. The lack of CERT protein suppresses the synthesis of SM [22].
The discovery and cloning of CERS1-6 have helped to understand the role of Cers with acyl chains of varying length in tumor cell signaling. For instance, CERS1 expression was found to be downregulated by histone deacetylase 1 (HDAC1) and miR5745p (microRNA, gene transcription regulation, cell-to-cell communication, vesicular transport) in the head and neck cancer cells, leading to the decrease in the content of Cer(C18:0). Cer(C18:0) produced by CERS1 induced cell death and suppressed tumor growth in cell cultures and xenografts of head and neck squamous cell carcinoma. About 50% mice lacking CERS2 activity necessary for the synthesis of long-chain Cer(C22-24) developed pheochromocytoma, presumably, due to the impaired apoptosis [23]. CERS6, which generates Cer(C16:0), was identified as a transcriptional target of p53. CERS6 was elevated in response to the transient expression of p53 or folate stress, which is known to activate p53, thus demonstrating that CERS6 is a component of the non-genotoxic p53-dependent cellular stress [24]. It also increased the susceptibility of colon cancer cells to apoptosis elicited by TRAIL (TNF-related apoptosis-inducing ligand) [25]. It was suggested that Cer(C16:0) influences the active transport (translocation) of caspase-3 into the nucleus or directly affects nuclear membrane permeability by increasing the nuclear pore size [25].
Cer(C16:0) also induced BAX-mediated apoptosis in irradiated HeLa37 cells. Mitochondrial anti-apoptotic Bcl2-like protein 13 (BCL2L13) directly inhibited Cer synthesis in cultured glioblastoma cells and tumor xenografts in mice by binding to CERS2 and CERS6 and blocking their homo- and heterodimerization [26].
Elevated levels of Cer(C16:0) generated by CERS6 were found in lung and oral cancer cells. Surprisingly, CERS6-generated Cer(C16:0) protected the integrity of the ER and Golgi membranes in HNSCC cells. Suppression of CERS6 affected delivery of the activating transcription factor 6 (ATF6) and caused its aberrant activation, leading to the ER stress-mediated death of HNSCC cells. Chen et al. [27] demonstrated a strong correlation between chemotherapy resistance in triple-negative breast cancer and upregulation of CERS6 expression.
Overall, these data confirm that CERS-generated Cers with acyl chains of different length play an important role in the death and survival of cancer cells, their exact effect being dependent on the tissue and type of cells [28, 29].
The total content of endogenous Cers was markedly elevated in malignant and benign breast tumors (12- and 4-fold, respectively) vs. normal tissues. This increase in the Cer content was rather unexpected, because it was demonstrated in in vitro studies that increased Cer levels were associated with the apoptosis induction, i.e., exhibited the anticarcinogenic effects [29]. Human head and neck squamous cell carcinomas were found to have an increased content of Cer(C16:0), Cer(C24:1), and Cer(C24:1) along with the decreased content of Cer(C18:0) [30].
Moreover, an increase in the Cer(C16:0) level due to the activity of CERS5 or CERS6 resulted in telomerase activation, while an increase in the content of Cer(C18:0) synthesized by CERS1 led to the inhibition of telomerase activity [24].
Biosynthesis of sphingomyelin. The ability of SM to promote tumor growth has been demonstrated in numerous studies. The presence of aberrantly high number of SM molecules in the apical layer of the surface membrane lowers its fluidity and permeability and increases membrane rigidity and durability. This interferes with the cell–cell communication, downregulates expression of cell surface molecules, and impairs coordination of signaling pathways, resulting in the activation of cell growth and uncontrolled proliferation, as well increase in the cell mobility and invasive potential [31]. The major signaling cascades involved in the intracellular effects of SM are RhoA/ROCK/LIMK/cofilin, RhoA/ROCK/FAK/paxillin [32], TGF-β/Smad, and EMT pathways [33].
Activation of metastasis due to a high SM content was first demonstrated in experiments with the metastatic prostate adenocarcinoma cell lines [34]. A possible mechanism of this effect might be related to the SM accumulation in the outer membrane that leads to the formation of additional hydrogen bonds with neighboring molecules and molecules of water followed by disruption of cell interaction with neighboring cells and extracellular matrix. The capacity to form a network of intermolecular hydrogen bonds is a feature of SM, which distinguishes it from, e.g., glycerolipids that contain ester groups that act as hydrogen bond acceptors, but not as donors [31].
SM generation is catalyzed sphingomyelin synthase (SMS). SMS exists as two isoforms (Table 3), one of which, SMS1, localizes to the trans-Golgi membranes, whereas SMS2 is associated with the plasma membrane. This distribution pattern accounts for the high SM concentration in the outer leaflet of the plasma membrane and in the trans-Golgi luminal membrane (Cer entering the cis-Golgi membrane is primarily utilized for the synthesis of GluCer rather than SM) [35].
Table 3. Human SMS isoforms [35, 36]
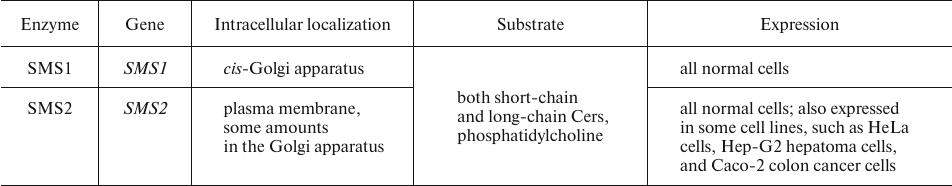
The impact of these two SMS isoforms on the SM production and their role in carcinogenesis remain controversial. Numerous studies have shown that proliferation and metastasis of tumor cell are mostly related to the activity of SMS2. For instance, Jing et al. [32] showed that a decline in the SMS2 activity suppressed migration, growth, and survival of ovarian cancer cells. Zheng et al. [33] reported upregulated expression of SMS2 in metastatic breast cancer.
However, SMS1 also plays a role in carcinogenesis. Van der Luit et al. [37] demonstrated that low SMS1 activity in S49 mouse lymphoma cells resulted in the decreased SM content in lipid rafts and abrogated cell apoptosis induced by alkyl-lysophospholipids.
On the other hand, Tafesse et al. [36] showed that SMS1 and SMS2 were co-expressed in various cell types, as well as demonstrated that both enzymes were equally necessary for the SM synthesis in cervical carcinoma cells. Downregulation of either SMS1 or SMS2 by RNA interference resulted in a profound decline in the SM production, Cer accumulation, and inhibition of cell growth. Moreover, it was observed that although SMS-deficient cells contained less SM, exposure to exogenous SM did not restore the growth of cells. Hence, these data evidence that the biological role of SMS goes beyond SM formation.
The activity of SMS in tumor cells is closely related to the cell sensitivity to chemotherapy. In particular, SMS2 inhibition substantially potentiated the efficacy of cisplatin in tumor treatment [32]. Doxorubicin-resistant vs. non-resistant HL-60/ADR leukemia cells demonstrated markedly elevated SMS activity [38].
Biosynthesis of cerebrosides. Cerebrosides also promote tumor growth. Human cells synthesize two types of cerebrosides (monoglycosylceramides): GalCer and GluCer.
A shift in the balance between Cers and their glycosylated counterparts towards the latter is caused, among other factors, by alterations in the activity of enzymes involved in their synthesis, which underlies the rise in the proliferative and invasive activity of tumor cells [39, 40] and accounts for their chemoresistance to some drugs [39]. For instance, the content of GalCer on the surface of many tumor cells is increased, resulting in the reduced cell adhesion and inhibition of apoptosis followed by the activation of cell proliferation and migration, increased cell survival, and promotion of carcinogenesis [39].
Similar to other sphingolipids, Cers can be involved in the regulation of tumor cell proliferation and metastasis via the Wnt and β-catenin pathway [41]. In particular, GluCer upregulates expression of the multidrug resistance protein 1 (MDR1) by activating nuclear transport of β-catenin and inhibiting its degradation [42]. Interestingly, GalCer plays a role in the transmembrane signaling by altering intracellular calcium levels. Joshi and Mishra [43] demonstrated that exposure to anti-GalCer antibodies resulted in the elevation of the intracellular calcium concentration caused by the Ca2+ influx through the plasma membrane and release from intracellular depots.
Reactions of GalCer synthesis catalyzed by galactosylcerebroside synthase (GalCERS) proceed in the ER. The highest expression of GalCERS was found in oligodendrocytes, Schwann cells, kidney epithelial cells, and testicles. The synthesis of GluCer by glucosylcerebroside synthase (GluCERS) takes place on the Golgi apparatus membrane [44].
Cabot et al. [45] demonstrated that the content of GluCer was elevated in several multidrug-resistant cell lines (MCF-7-AdrR breast cancer, KB-V-1 vinblastine-resistant epidermoid carcinoma, and OVCAR-3 ovarian adenocarcinoma cells). It was suggested that it was related to the activation of GluCer synthesis, because the rate of GluCer degradation remained unaltered. A comparable increase in the GluCer content was also reported for chemotherapy-resistant human tumors [46]. Upregulation of GluCERS expression in MCF-7 breast cancer and leukemia cells made these cells resistant to chemotherapy [38, 45]. GalCERS activity is increased in tumor cells; it is also higher in chemotherapy-resistant vs. chemotherapy-sensitive cells. Thus, Itoh et al. [38] reported a 2-fold increase in the GalCERS activity in doxorubicin-resistant vs. nonresistant HL-60/ADR leukemia cells.
Biosynthesis of gangliosides. Some gangliosides regulate signal transduction mediated by receptor tyrosine kinases (RTKs) [47]. Typically, they form complexes with RTKs in lipid rafts and contribute to the activation/inhibition of signaling via these receptors. Changes in the ganglioside levels in the plasma membrane affect molecular composition and structure of lipid rafts resulting in the RTK reorganization and/or exclusion from the lipid rafts [48].
Localization of the CD95 death receptor to the lipid rafts is essential for efficient apoptosis signaling. The structure and composition of ganglioside-containing rafts play a decisive role in the regulation of cell death and survival [48]. Using lymphoid and myeloid tumor cell models, it was found that gangliosides trigger apoptosis by activating CD95 and accumulating in the mitochondrial membrane, which disturbs mitochondrial transmembrane potential and induces apoptosis via a caspase-independent pathway [49].
Veldman et al. [50] showed that multidrug-resistant tumor cells had a reduced content of lactosylcerebrosides and gangliosides together with the elevated levels of GluCer, GalCer, and SM. It was suggested that the glycolipid biosynthesis pathway in these cells is arrested at the level of lactocerebroside formation leading to the GluCer accumulation.
The role of gangliosides and enzymes involved in their synthesis/degradation in the regulation of tumor cell death and survival has been discussed in detail in a recent review by Sasaki et al. [48], so we will describe it only briefly.
Biosynthesis of sulfatides. Sulfatides are produced by cerebroside sulfotransferase (CST) (encoded by the GAL3ST1 gene) in the lumen of the Golgi apparatus. De novo synthesized plasma membrane and sulfatides synthesized in the ER, as well as “aged” sulfatides, are redistributed within the cell by vesicular transport involving clathrin or carrier proteins, such as glycolipid transfer protein (GLTP). Besides being directly involved in sulfatide transport, GLTP acts as a sensor of glycolipid levels in the cell [51].
Sulfatides are synthesized in many types of cells. The highest content of these compounds (4-7% of total lipids) was found within the myelin sheaths formed by oligodendrocytes and Schwann cells in the central and peripheral nervous system, respectively. Sulfatides are also found in other glial cells, such as astrocytes (which produce them in small amounts) and in neurons (which sulfatides enter by endocytosis). Large amounts of sulfatides are typical for human kidneys, gastric and duodenal mucosa, cells of the islet of Langerhans, and membranes of red blood cells, platelets, and granulocytes [52].
An increase in the sulfatide content and upregulation of CST expression have been found in many tumor types. For instance, high CST expression and activity in the absence of changes in the arylsulfatase A (ARSA) activity resulted in the increased content of sulfated glycolipids in kidney cancer vs. normal cells. Moreover, an increased ARS activity was found in the serum of patients with renal cell carcinoma and hepatocellular carcinoma compared to the healthy subjects. However, unlike in renal cell carcinoma, the high levels of CST activity in the serum of patients with hepatocellular carcinoma did not correlate with the level of enzyme activity in the tumor and were similar to those observed in normal tissue. In the case of gastric cancer, CST expression varied in both patients and healthy subjects [52, 53]. In SMKT-R3 human kidney cancer cells, the rise in the sulfotransferase activity was driven by the tumor necrosis factor α (TNF-α). It was found that the secreted form of TNF-α binds to the cognate receptor, while membrane-bound TNF-α provides direct contact between neighboring cells. The activity of CST in kidney carcinoma cells increased after exposure to the epidermal growth factor (EGF) [53, 54].
An increase in the content of sulfatides has been detected in low-grade endometrial adenocarcinoma, some lung tumors, brain and colon tumors, hepatocellular carcinoma, and ovarian cancer. The use of sulfatides as early markers in the diagnostics of ovarian cancer has been suggested in several studies. Thus, mass spectrometry analysis of cell metabolites along with the assessment of gene expression profiles revealed an increase in the content of sulfatides in ovarian cancer cells vs. normal tissue. This observation was confirmed by the fact that ovarian epithelial carcinoma cells demonstrated higher levels of mRNAs encoding GalCERS and CST necessary for sulfatide synthesis, whereas the levels of ARS, saposin, and galactosylceramidase (GalCERase) remain the same [51].
Six human kidney carcinoma cell lines (SMKT-R1, SMKT-R2, SMKT-R3, SMKT-R4, TOS-1, and TOS-2) were found to have a markedly elevated CST activity and higher content of CST mRNA, whereas the increase in the content of this mRNA in ACHN renal cancer vs. normal cells was only modest. At the same time, no rise in the sulfatide level was observed in the case of Wilms tumor (nephroblastoma). Hence, overproduction of sulfatides is not necessarily typical for all cancer types. Furthermore, it still remains to be clarified whether elevated sulfatide levels are an artifact observed in cultured tumor cells (since lipids can migrate between the cell membrane and serum-containing medium) or they represent an in vivo phenomenon [54].
Biosynthesis of ceramide-1-phosphate. Ceramide-1-phosphate (C1P) is a signaling molecule that acts antagonistically to Cer. Similarly to sphingosine-1-phosphate (S1P), it is a potent apoptosis inhibitor that promotes cell survival, DNA synthesis, and cell division.
C1P blocks apoptosis by inhibiting either acidic sphingomyelinase (aSMase) or caspase, as well as by preventing DNA fragmentation in macrophages. It also inhibits serine palmitoyltransferase, a key regulatory enzyme in the biosynthesis of long-chain sphingoid bases and, therefore, Cers. C1P is released by damaged cells; it is also found in exosomes exported to the plasma and is able to interact with the plasma membrane of other cells at specific sites [55].
The mitogenic effect of C1P is mediated by multiple signaling pathways and enzymes, including MEK/ERK1-2, NADPH oxidases, protein kinase C, and PI3K/Akt/mTOR. Beside activating cell proliferation, C1P promotes cell survival by inhibiting apoptosis-related signaling pathways. It stimulates the PI3K/Akt-axis and upregulates expression of inducible NO synthase (iNOS) [8].
C1P is synthesized from Cer by ceramide kinase (CERK). CERK can associate with various membranes, including those most distant from the nucleus, such as membranes of trans-Golgi, endosomes, and mitochondria, as well as the plasma membrane. CERK is activated by interleukin-1β and calcium ions. Generated C1P activates type IVA phospholipase A2 (cPLA2) in the Golgi apparatus membrane and promotes release of arachidonic acid, thus stimulating the synthesis of pro-inflammatory eicosanoids [56].
Different types of cells can differ in the subcellular CERK distribution and expression. CERK is regulated by phosphorylation/dephosphorylation and cations (mainly Ca2) along with translocation between intracellular compartments [57].
Multiple studies have demonstrated high CERK activity in breast [58] and pancreatic cancer [59]. In breast tumors, upregulation of CERK expression correlates with the risk of disease recurrence [60].
Biosynthesis of sphingosine-1-phosphate. S1P regulates cell survival and proliferation [61]. The biological effects of S1P are realized via two mechanisms: (i) S1P export from the cell and exertion of its paracrine (or autocrine) action, or (ii) binding to intracellular targets [62]. S1P is a charged lipid that cannot freely diffuse across the membrane. It is transported by low-selective ATP-binding transport proteins of the ABC family, as well as highly selective SPNS2 protein of the MFS family [63]. SPNS2 transports both S1P and dihydrosphingosine-1-phosphate (dhS1P), but not their non-phosphorylated counterparts. The transport of S1P by SPNS2 increases proportionately to the increase in the intracellular S1P level, suggesting that SPNS2 acts as a passive transporter that requires no energy for its activity, which distinguishes it from the ABC transporters [64].
The concentration of S1P in the blood plasma is maintained at 0.5-1 μM due to the production by erythrocytes, endothelial cells, and liver cells. Liver cells synthesize ApoM, a physiological S1P carrier that transports it to the high-density lipoproteins (HDLs). The concentration of S1P in the interstitial fluid is maintained at a low nanomolar level due to high activity of S1P-degrading enzymes (e.g., sphingosine-1-phosphate lyase S1PL) [65].
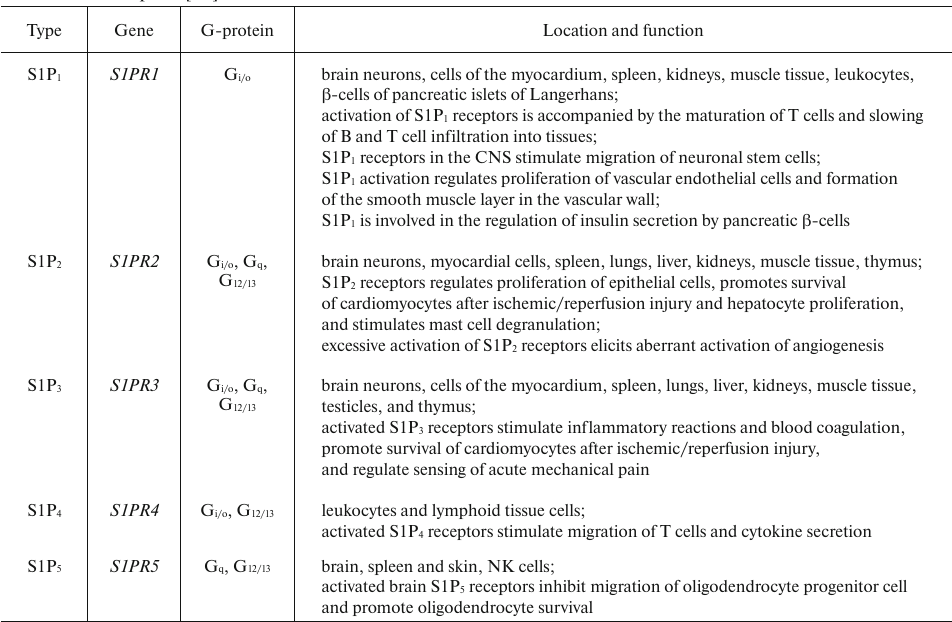
S1P binds to five membrane receptors (S1PRs) belonging to the GPCR superfamily (Table 4). S1PR-activated signaling pathways regulate the development of CNS and cardiovascular system, reproductive function, migration of immune cells, adhesion, cell survival and division, stress response, etc. S1P also regulates cellular response by directly binding to some intracellular targets, such as HDACs [66]. Similar to the vascular endothelial growth factor, S1P increases vascular permeability by activating S1P2 receptor and Rho/ROCK/PTEN signaling [67]. S1P can also prevent apoptosis by inhibiting translocation of cytochrome c and Smac/DIABLO from the mitochondria [68].
S1P affects metastasis, cell migration, angiogenesis and lymphogenesis. S1P molecules circulating in the bloodstream and produced by the tumor are involved in the activation of angiogenesis and lymphogenesis. Ponnusamy et al. [69] reported that S1P neutralization in the bloodstream by specific antibodies suppressed metastasis development in the lungs. S1P induced metastasis of liver carcinoma cells by establishing the MMP-7/syndecan-1/TGF-β1 autocrine loop [1]. By activating ERK, AKT, and p38, S1P stimulates chemotaxis and receptor-dependent invasion by ovarian cancer cells [70]. S1P regulates cell migration potential by redistributing actin from focal junctions to membrane folds (lamellipodia). In MCF-7 cells, S1P stimulated actin redistribution in membrane folds and promoted cell migration, whereas a decline in the S1P content restored the contacts in the actin-containing adhesion foci and prevented cell migration [66]. S1P stimulated migration of MDA-MB-231 breast cancer cells and potentiated their metastatic activity [71].
S1P is directly synthesized from sphingosine (Sph) by sphingosine kinases (SPHKs) that transfers phosphate from ATP molecule to the C1 hydroxyl group of Sph. As described below, Sph is generated in the SM catabolism mostly due to the activity of SMase and ceramidase (CERase) [44]. Two human genes, SPHK1 and SPHK2, code for SPHK1 and SPHK2, respectively. Each of these enzymes has several splice variants. Although SPHK1 and SPHK2 significantly differ in size, they share a high degree of sequence similarity. Almost entire SPHK1 polypeptide is homologous to the fragments of larger SPHK1 molecule, but the N-termini of the two proteins differ. SPHK2 also has a proline-rich segment in the central portion of the protein that is not found in SPHK1 or any other known protein [72]. The characteristics of human SPHK isoforms are presented in Table 5.
Table 5. Human SPHK isoforms [73-76]
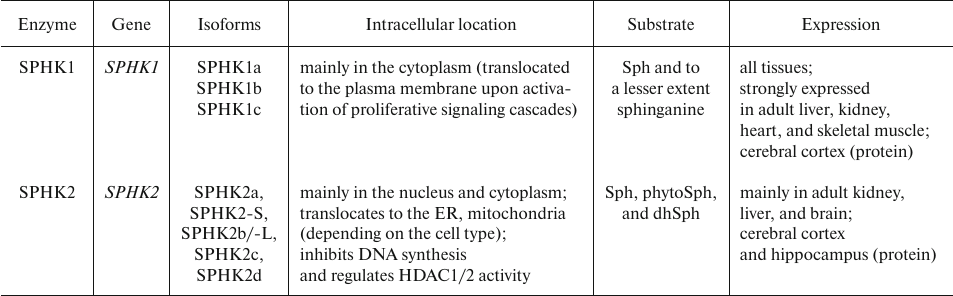
Note. Enzyme isoforms are designated in accordance with GenBank and
Uniprot. The data on the number (5 or 7) of SPHK2 isoforms differ, as
not all of them have been confirmed experimentally.
An increase in the SPHK1 activity accounting for the elevated S1P level has been observed in many tumors. For instance, upregulated expression of SPHK1 mRNA and/or SPHK1 protein was found in gastric, lung, brain, and colon cancers [77]. Metastatic vs. non-metastatic colon cancer is characterized by the elevated SPHK1 expression [78]. High levels of SPHK1 expression in tumors also correlate with poor survival and induced tamoxifen resistance in patients with estrogen-sensitive breast cancer [66].
The effect of SPHK2 activity is more complex. Early studies have demonstrated that SPHK2 overexpression results in the suppression of cell growth and apoptosis activation, suggesting that although both SPHK2 and SPHK1 utilize similar physiological substrates and generate the same final product, SPHK2 plays the opposite role to that of SPHK1. The pro-apoptotic effect of SPHK2 has been supported by more recent studies demonstrating that mouse mesangial Sphk2–/– cells are more resistant to staurosporine-induced apoptosis compared to the wild-type or Sphk1–/– cells. Moreover, downregulation of endogenous SPHK2 expression in HEK293 cells led to the arrest of the TNF-α-induced apoptosis. However, SPHK2 can also play the anti-apoptotic role, as suppression of SPHK2 expression or SPHK2 activity with isoform-specific inhibitors promoted apoptosis and increased the sensitivity of tumor cells to chemotherapy [72].
RELATION BETWEEN THE ACTIVITY OF SPHINGOLIPID DEGRADATION ENZYMES
AND TUMOR PROGRESSION
Cerebroside degradation. Cerebroside hydrolysis catalyzed by galactosylceramidase (GalCERase) and glucosylceramidase (GluCERase), respectively, results in the formation of ceramides and corresponding monosaccharides (galactose or glucose) [44].
Several studies have demonstrated a decline in the GalCERase activity in tumors, in particular, due to the decrease in the GALC gene expression. For example, GALC expression is downregulated in the head and neck [39, 79], lung [79], and nasopharyngeal [80] cancers. Zhao et al. [80] showed that upregulated GALC expression in CNE-2Z cells suppressed cell proliferation, migration, and metastatic potential.
Downregulation of GALC transcription can result from the binding of inhibitory transcription factors to the gene promoter region. The GALC gene is located on chromosome 14q3 and contains sequences interacting with the transcription factors SP1 (5′-CCCGCC-3′), YY1 (5′-AAATGG-3′), and AP2 (5′-GCCTGCAGGC-3′) [81]. Among these transcription factors, the most probable inhibitor is YY1, whose role in the transcription suppression has been described earlier [39]. For instance, Sui et al. [82] and Yakovleva et al. [83] demonstrated the inhibitory effect of YY1 on the transcription of p53 gene. Another potential mechanism for the suppression of GALC transcription is hypermethylation of its promoter [79].
Recent studies have shown that GalCERase can exert the opposite effect on tumor growth and cell differentiation, which casts doubt on its impact on sphingolipid metabolism in cancer cells, as well as on its role in tumor progression [84, 85]. In particular, Belleri et al. [85] showed that GalCERase inhibition in mouse B16-F10 and human A2058 melanoma cells resulted in the activation of the SMPD3 gene coding for SMase, an enzyme involved in Cer generation. This ultimately increased the level of Cer and suppressed the growth, motility, and invasive ability of melanoma cells, as well as inhibited the metastatic activity of B16-F10 cells grafted in syngeneic mice or Danio rerio embryos. GalCERase triggered the opposite effect. These data were corroborated by the fact that progressively upregulated GALC expression in human skin samples, ranging from common nevus to stage IV melanoma, was accompanied by proportional downregulation of the SMPD3 expression and decrease in the Cer level.
The impact of GluCERase on tumor cell growth, proliferation, and metastasis is remarkedly less pronounced than that of GalCERase (Table 6).
Table 6. Human GluCERase isoforms [86]
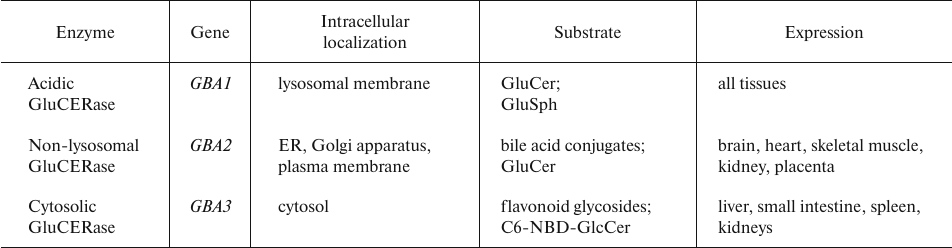
It has been noticed that the development of Gaucher disease caused by the GluCERase deficiency correlates with the emergence of multiple myeloma, leukemia, glioblastoma, lung cancer, and hepatocellular carcinoma, although the underlying cause for this association is poorly understood. However, only few researchers believe that the development of these tumors resulted from the GluCERase deficiency; most scientists consider their emergence to be related to the therapy of Gaucher disease [86].
Sphingomyelin degradation. The key enzymes in sphingomyelin catabolism, sphingomyelinases (SMases), belong to the family of phosphodiesterases, whose catalytic mechanism is similar to that of phospholipases C. SMases transform SM into Cer and phosphocholine. Several isoforms of human SMase differing in their pH optimum and, therefore, cellular location have been characterized. There are also Mg2+- and Zn2+-dependent forms of the enzyme, as well as the isoform independent of these ions (Table 7) [44, 87, 88].
Table 7. Human SMase isoforms [87-93]
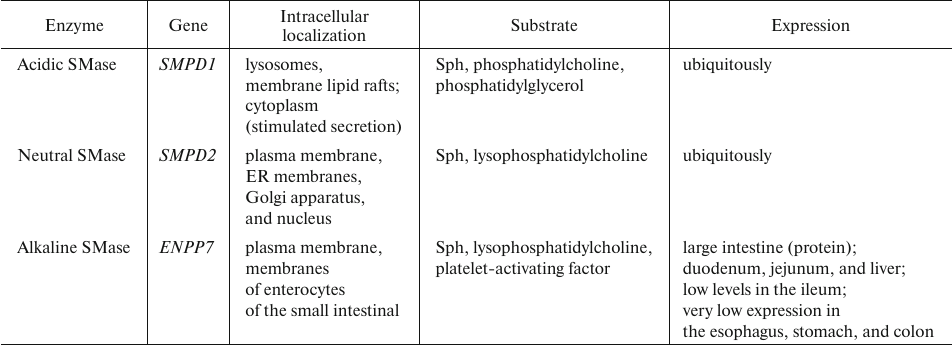
The antitumor activity of SMase is related to its role in the generation of Cer, which suppresses proliferation of tumor cells. High SMase activity results in the elevated Cer levels and vice versa. Changes in the SMase activity also affect the structure and function of membranes. About 70% total cellular SMase is located in lipid rafts. Degradation of plasma membrane sphingomyelins by SMase increases membrane fluidity and affects cholesterol distribution in the cell, in particular, displacement of cholesterol with Cer in the membrane lipid rafts [87, 94]. Platelet-derived acidic SMase stimulates cell adhesion and melanoma metastasis. Furthermore, it was assumed that in endothelial cells, SMase plays a key role in tumor angiogenesis, because the SMase-induced increase in Cer levels was shown to mediate cell sensitivity to the anti-angiogenic therapy [95].
As early as in 1994, SMase was proven to take part in the apoptotic death of tumor cells when Jarvis et al. [96] reported that the TNF-α-induced SMase activation resulted in the Cer-induced apoptosis in HL-60 and U937 human leukemic cells and in L929/LM and WEHI 164/13 mouse fibrosarcoma cells.
The activity of SMase is decreased in tumor cells. Epigenetic analysis revealed that the gene for neutral SMase (nSMase) was hypermethylated and, therefore, its expression was downregulated in breast cancer [89], MDA-MB-231 triple negative breast cancer [87], and hepatocellular carcinoma [91] cell lines. In contrast, no nSMase gene hypermethylation was found in MCF7 cells, but the activity of the enzyme was low, suggesting that expression of its gene was regulated by a different mechanism, e.g., via a direct effect on histone acetylation [90]. It was also indirectly proven that the activity SMase is altered in tumor cells. For example, Kim et al. [92] identified point mutations in the gene encoding nSMase2 in 5 and 6% samples of acute myeloid leukemia and acute lymphoblastic leukemia, respectively. Although functional implications of the majority of these mutations remain unclear, two of them were found to decreased the enzyme stability and cause its aberrant localization. The authors also discovered a deletion in the nSMase-encoding gene in mouse osteosarcoma F4328 cells.
The mechanism of action of some anticancer agents involves SMase activation. Aslan et al. [97] showed that thymoquinone induced SMase-mediated apoptosis in MCF-7 breast cancer and HepG2 cells via activation of nSMase and the following rise in the Cer levels. Kilkus et al. [98] suggested a similar mechanism of nSMase activation-dependent Cer-induced apoptosis in staurosporine-exposed LA-N-5 human neuroblastoma cells. Rotolo et al. [99] showed that exposure of Jurkat cells to UV radiation resulted in the translocation of secreted aSMase to the lipid microdomain and assembly of Cer-rich membrane platforms, resulting in the activation of apoptosis. Ferranti et al. [100] demonstrated that the UV-induced aSMase activation and secretion occurred via the calcium-dependent fusion of lysosomes and plasma membrane. Clarke et al [101] showed that the antitumor effect of doxorubicin was associated with the activation of the SMPD2 gene promoter, which upregulated enzyme synthesis and increased the content of Cers. SMase activity depends on multiple factors and can be potentiated by TNF-α, 1,25-dihydroxycalciferol, interferon gamma (IFNγ), nerve growth factor, heat, ionizing radiation, fenretinide, and arabinosylcytosine [45, 102].
Ceramide degradation. Excessive Cer amounts pose a threat to the cell, so Cers are rapidly hydrolyzed by CERase (Table 8) into free amino alcohols (Sph and its analogs) and fatty acids. Free intracellular Sph is produced solely by the CERase-mediated Cer catabolism. Like other sphingoid bases, Sph can be released from the lysosomes and reutilized in the CERS-mediated Cer synthesis or phosphorylated with the formation of S1P [44, 103, 104].
A decline in the endogenous Cer level and activation of S1P synthesis due to the high CERase activity shift the sphingolipid balance towards a state promoting cell survival [105].
Table 8. Human CERase isoforms [103, 106]
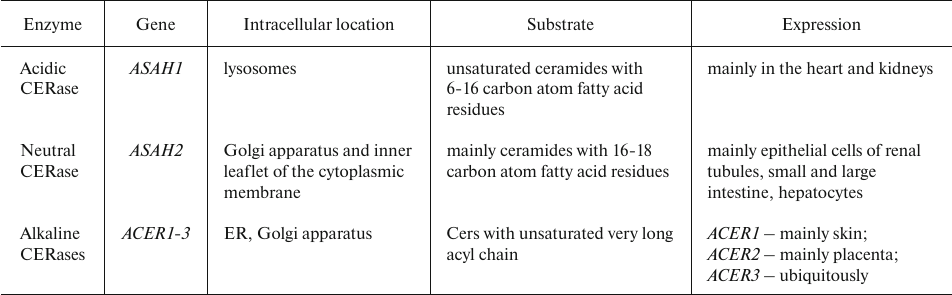
Acidic CERase (aCERase) plays a major role in cancer development and progression. It is highly expressed in solid prostate and breast cancers, melanoma, and leukemia [105]. At the same time, it was shown that the content of aCERase was decreased in thyroid tumor cells [106].
The high activity of aCERase followed by a decline in the Cer content may underlie resistance of tumors to chemotherapy. Thus, a decrease in the Cer level due to the ASAH1 overexpression was found to be associated with resistance to cisplatin [107].
Sulfatide degradation is mediated by lysosomal arylsulfatase A (ARSA). Human ARS family includes 11 members (A-K isoforms). Human ARSA and ARSB localize to the lysosomes; ARSC is a microsomal protein, whereas ARSD, F, H, J, and K are located in the ER membrane. ARSE is found in the Golgi apparatus. ARSG was detected in both ER and lysosomes. ARSI is typical to the ER but can also be secreted to the extracellular environment. All family members share 20-60% sequence homology [108].
The activity of ARSA depends on saposin B (SapB) that extracts sulfatide moieties from membranes thereby making them available to the enzyme. An alternative sulfatase-independent pathway for sulfatide degradation was found in neuroblastoma cell line, in which Cer was directly produced from sulfatide without prior desulfation [109].
Sulfatides accumulated due to the ARSA deficiency or mutations in the SapB-encoding gene are associated with demyelination and lethal neurological disease metachromatic leukodystrophy. Overall, the available data suggest that impairments in the sulfatide production have a significant effect on the emergence of neuronal defects [51].
ARS has been actively used in experiments on the role of sulfatides in metastasis. In particular, in vitro adhesion of activated platelets expressing P-selectin to MC38 mouse colon cancer cells solely involved sulfatides produced by the latter cells. Interaction between MC38 cells and platelets was observed in vivo after transplantation of MC38 cells into mice; the aggregates of tumor cells with platelets were found as early as 30 min after intravenous injection of tumor cells. In contrast, removal of sulfate groups by ARS resulted in the loss of metastatic potential of MC38 cells, thus evidencing that sulfatides directly govern formation of colon cancer cell aggregates [53].
ARSA can be used for desulfation. Thus, metabolic inhibition of sulfation by treatment of MC38 cells with ARSA impaired the binding of P-selectin, which, in turn, suppressed development of metastases in mouse lungs. Sulfatides that are present in high amounts on the surface of tumor cells can presumably bind ligate P-selectin, thus promoting the spreading of the tumor. However, the relationship between the tumor progression and elevated expression of sulfatides remains unclear [110, 111].
Ceramide 1-phosphate degradation. C1P dephosphorylation involves several phosphatases. For example, the phosphate group clan be cleaved from C1P by C1P phosphatase with the formation of Cer. C1P phosphatase, which is present in high amounts in the brain synaptosomes and plasma membrane of liver cells, differs from PA phosphohydrolase that hydrolyzes phosphatidic acid (PA). Nevertheless, PA phosphohydrolase, which is located in the plasma membrane, can also convert C1P into Cer. This enzyme belongs to a family containing at least three mammalian phosphatases (lipid phosphate phosphatases, LPPs) that degrade various lipid phosphates, LPPs regulate cell survival by controlling the intracellular levels of PA and S1P, as well as leukocyte infiltration and airway inflammation. C1P dephosphorylation attenuate/inactivate its regulatory effects, although the resulting Cer can be harmful to the cells. The balance between the content of Cer and C1P coordinated by CERK and C1P phosphatase in important for the functioning of metabolic or signaling pathways regulated by these two sphingolipids [112-115].
Sphingosine 1-phosphate degradation. Intracellular S1P catabolism is catalyzed by sphingosine 1-phosphate phosphatases (SGPPs) and S1PLs (Table 9) [116].
Highly homologous SGPPs form the SGPP family, the members of which display a rather narrow substrate specificity and dephosphorylate solely S1P, dhS1P, and phytoS1P. They are integral ER membrane proteins involved in the regulation of intracellular S1P level and can replenish the cellular pool of free Sph [44, 116].
Table 9. Essential enzymes involved in S1P
degradation [44, 116]
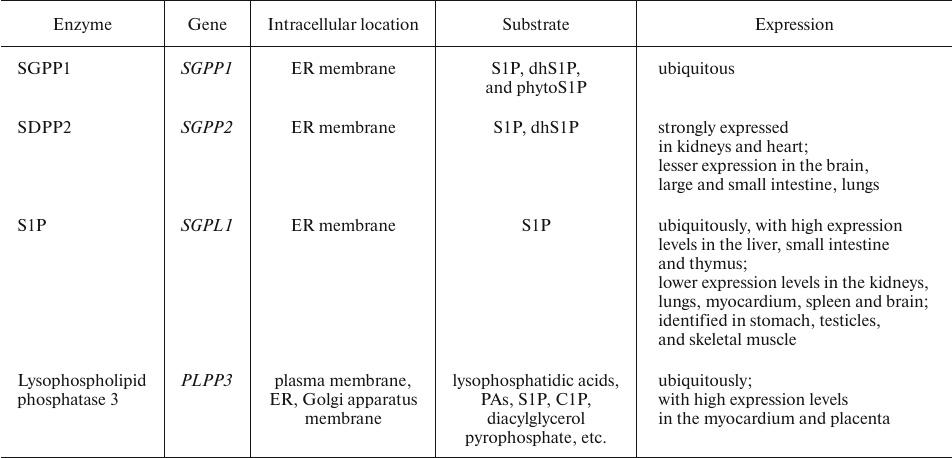
Excessive blood plasma S1P is cleaved on the hepatocyte plasma membrane by lysophospholipid phosphatase 3, an enzyme of the LPP family [44, 116].
S1PL can also convert S1P into ethanolamine-1-phosphate and C16 aldehyde (hexadecenal). In live cells, this reaction is irreversible and therefore, represents an intracellular mechanism for sphingolipid elimination [42].
S1PL is involved in the in both GPR55-dependent and independent pathways of cell proliferation [117]. Although S1PL expression S1Plase is altered in human tumor tissues, the role of S1PL in cancer pathogenesis has not yet been fully elucidated [117]. Thus, the activity of S1PL is increased in many types of tumor cells, e.g., in ovarian cancer [118]. However, the levels of S1PL remained unaltered or even decreased in hepatocellular carcinoma [119] and colon cancer [120] cells, despite an increase in the SPHK. Most likely, this was due to the increase in the S1PL activity involved in S1P utilization [117]. Other studies demonstrated that the S1PL activity was decreased in colon cancer [121].
CONCLUSION
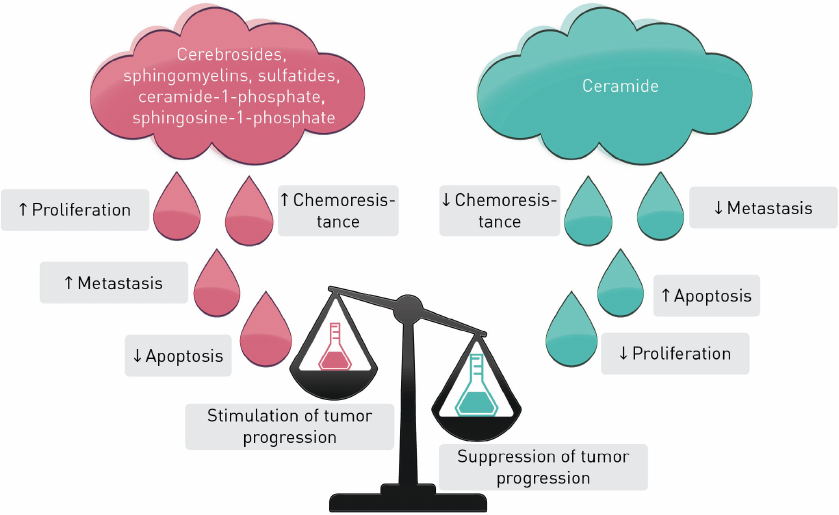
S1P, C1P, Cer, dhCer, and other sphingolipids are the major players in sphingolipid cycle, the dysregulation of which can elicit cell death or uncontrolled cell proliferation (Fig. 3). Typically, Cers cause cell cycle arrest or cell death in response to cell stress whereas S1P, C1P, sphingomyelins, cerebrosides, and sulfatides promote cell survival and proliferation. Interconversion of these metabolites catalyzed by a set of enzymes provides an opportunity for a fine regulation of the cellular processes.
Fig. 3. Stimulation and suppression of tumor progression by sphingolipids.
Based on the analyzed data, we can formulate the following main features of sphingolipid metabolism in tumor cells.
- Generally, tumor cells are characterized by the decreased level of enzymes involved in Cer production, low activity of these enzymes, and content of phosphorylated Cer, Sph, and their derivatives, which promotes proliferation and survival of tumor cells.
- Increased content of sulfatides on the cell membrane surface contributes to metastasis development and spread in some tumor cell types.
- The majority of tumor cells is characterized by increased S1P and C1P levels; high concentrations of these compounds promote cell proliferation and metastasis. In the case of S1P, this effect is mediated largely due to the activation of angiogenesis.
- The shift of the dhCer/Cer balance toward dhCer activates cell proliferation and metastasis in various tumor types. The shift of the balance toward Cer exerts the opposite effects.
- Alterations in the cell membrane composition due to the degradation of common lipids in tumor cells leads to the rearrangement of membrane fragments, including lipid rafts, resulting in the “elimination” of specific membrane receptors, which inhibits apoptosis and activates cell proliferation.
Contributions. V.S.P. conceived and supervised the study and edited the manuscript; V.I.I.-R. and O.M.K. wrote and edited the manuscript.
Funding. The study was carried out within the framework of the State Task of the RUDN University of the Ministry of Science and Higher Education no. 075-01551-23-00 (FSSF-2023-0006).
Ethics declarations. The authors declare no conflict of interests. This article does not contain description of studies with the involvement of humans or animal subjects performed by any of the authors.
REFERENCES
1.Li, R. Z., Wang, X. R., Wang, J., Xie, C., Wang, X.
X., Pan, H. D., Meng, W. Y., Liang, T. L., Li, J. X., Yan, P. Y., Wu,
Q. B., Liu, L., Yao, X. J., and Leung, E. L. (2022) The key role of
Sphingolipid metabolism in cancer: New therapeutic targets, diagnostic
and prognostic values, and anti-tumor immunotherapy resistance,
Front. Oncol., 27, 941643, doi:
10.3389/fonc.2022.941643.
2.Lin, M., Li, Y., Wang, S., Cao, B., Li, C., and Li,
G. (2022) Sphingolipid metabolism and signaling in lung cancer: a
potential therapeutic target, J. Oncol., 28, 9099612,
doi: 10.1155/2022/9099612.
3.Pal, P., Atilla-Gokcumen, G. E., and Frasor, J.
(2022) Emerging roles of ceramides in breast cancer biology and
therapy, Int. J. Mol. Sci., 23, 11178, doi:
10.3390/ijms231911178.
4.Pherez-Farah, A., López-Sánchez, R.
D. C., Villela-Martínez, L. M., Ortiz-López, R.,
Beltrán, B. E., and Hernández-Hernández, J. A.
(2022) Sphingolipids and lymphomas: a double-edged sword, Cancers
(Basel), 14, 2051, doi: 10.3390/cancers14092051.
5.Byrne, F. L., Olzomer, E. M., Lolies, N., Hoehn, K.
L., and Wegner, M. S. (2022) Update on glycosphingolipids abundance in
hepatocellular carcinoma, Int. J. Mol. Sci., 23, 4477,
doi: 10.3390/ijms23094477.
6.Zaibaq, F., Dowdy, T., and Larion, M. (2022)
Targeting the sphingolipid rheostat in gliomas, Int. J. Mol.
Sci., 23, 9255, doi: 10.3390/ijms23169255.
7.Sukocheva, O. A., Furuya, H., Ng, M. L.,
Friedemann, M., Menschikowski, M., Tarasov, V. V., Chubarev, V. N.,
Klochkov, S. G., Neganova, M. E., Mangoni, A. A., Aliev, G., and
Bishayee, A. (2020) Sphingosine kinase and sphingosine-1-phosphate
receptor signaling pathway in inflammatory gastrointestinal disease and
cancers: a novel therapeutic target, Pharmacol. Ther.,
207, 107464, doi: 10.1016/j.pharmthera.2019.107464.
8.Camacho, L., Ouro, A., Gomez-Larrauri, A.,
Carracedo, A., and Gomez-Muñoz, A. (2022) Implication of
ceramide kinase/C1P in cancer development and progression, Cancers
(Basel), 14, 227, doi: 10.3390/cancers14010227.
9.Belleri, M., Chiodelli, P., Corli, M., Capra, M.,
and Presta, M. (2022) Oncosuppressive and oncogenic activity of the
sphingolipid-metabolizing enzyme β-galactosylceramidase,
Biochim. Biophys. Acta Rev. Cancer, 1877, 188675, doi:
10.1016/j.bbcan.2021.188675.
10.Ogretmen, B. (2017) Sphingolipid metabolism in
cancer signalling and therapy, Nat. Rev. Cancer, 18,
33-50, doi: 10.1038/nrc.2017.96.
11.Zelnik, I. D., Rozman, B., Rosenfeld-Gur, E.,
Ben-Dor, S., and Futerman, A. H. (2019) A stroll down the CerS lane,
Adv. Exp. Med. Biol., 1159, 49-63, doi:
10.1007/978-3-030-21162-2_4.
12.Stiban, J., Tidhar, R., and Futerman, A. H.
(2010) Ceramide synthases: roles in cell physiology and signaling,
Adv. Exp. Med. Biol., 688, 60-71, doi:
10.1007/978-1-4419-6741-1_4.
13.Stiban, J., Fistere, D., and Colombini, M. (2006)
Dihydroceramide hinders ceramide channel formation: implications on
apoptosis, Apoptosis, 11, 773-780, doi:
10.1007/s10495-006-5882-8.
14.Lachkar, F., Ferré, P., Foufelle, F., and
Papaioannou, A. (2021) Dihydroceramides: their emerging physiological
roles and functions in cancer and metabolic diseases, Am. J.
Physiol. Endocrinol. Metab., 320, E122-E130, doi:
10.1152/ajpendo.00330.2020.
15.Zheng, W., Kollmeyer, J., Symolon, H., Momin, A.,
Munter, E., Wang, E., Kelly, S., Allegood, J. C., Liu, Y., Peng, Q.,
Ramaraju, H., Sullards, M. C., Cabot, M., and Merrill, A. H., Jr.
(2006) Ceramides and other bioactive sphingolipid backbones in health
and disease: lipidomic analysis, metabolism and roles in membrane
structure, dynamics, signaling and autophagy, Biochim. Biophys.
Acta, 1758, 1864-1884, doi:
10.1016/j.bbamem.2006.08.009.
16.Breen, P., Joseph, N., Thompson, K., Kraveka, J.
M., Gudz, T. I., Li, L., Rahmaniyan, M., Bielawski, J., Pierce, J. S.,
Van Buren, E., Bhatti, G., and Separovic, D. (2013) Dihydroceramide
desaturase knockdown impacts sphingolipids and apoptosis after
photodamage in human head and neck squamous carcinoma cells,
Anticancer Res., 33, 77-84.
17.Casasampere, M., Ordoñez, Y. F., Pou, A.,
and Casas, J. (2016) Inhibitors of dihydroceramide desaturase 1:
therapeutic agents and pharmacological tools to decipher the role of
dihydroceramides in cell biology, Chem. Phys. Lipids,
197, 33-44, doi: 10.1016/j.chemphyslip.2015.07.025.
18.Guo, W., Zhang, C., Feng, P., Li, M., Wang, X.,
Xia, Y., Chen, D., and Li, J. (2021) M6A methylation of DEGS2, a key
ceramide-synthesizing enzyme, is involved in colorectal cancer
progression through ceramide synthesis, Oncogene, 40,
5913-5924, doi: 10.1038/s41388-021-01987-z.
19.Hernández-Tiedra, S., Fabriàs, G.,
Dávila, D., Salanueva, Í. J., Casas, J., Montes, L. R.,
Antón, Z., García-Taboada, E., Salazar-Roa, M., Lorente,
M., Nylandsted, J., Armstrong, J., López-Valero, I., McKee, C.
S., Serrano-Puebla, A., García-López, R.,
González-Martínez, J., Abad, J. L., Hanada, K., Boya, P.,
and Velasco, G. (2016) Dihydroceramide accumulation mediates cytotoxic
autophagy of cancer cells via autolysosome destabilization,
Autophagy, 12, 2213-2229, doi:
10.1080/15548627.2016.1213927.
20.Grassme, H., Jekle, A., Riehle, A., Schwarz, H.,
Berger, J., Sandhoff, K., Kolesnick, R., and Gulbins, E. (2001) CD95
signaling via ceramide-rich membrane rafts, J. Biol. Chem.,
276, 20589-20596, doi: 10.1074/jbc.M101207200.
21.Crivelli, S. M., Giovagnoni, C., Zhu, Z.,
Tripathi, P., Elsherbini, A., Quadri, Z., Pu, J., Zhang, L., Ferko, B.,
Berkes, D., Spassieva, S. D., Martinez-Martinez, P., and Bieberich, E.
(2022) Function of ceramide transfer protein for biogenesis and
sphingolipid composition of extracellular vesicles, J. Extracell.
Vesicles, 11, e12233, doi: 10.1002/jev2.12233.
22.Giussani, P., Colleoni, T., Brioschi, L., Bassi,
R., Hanada, K., Tettamanti, G., Riboni, L., and Viani, P. (2008)
Ceramide traffic in C6 glioma cells: evidence for CERT-dependent and
independent transport from ER to the Golgi apparatus, Biochim.
Biophys. Acta, 1781, 40-51, doi:
10.1016/j.bbalip.2007.11.002.
23.Chen, W., Wu, C., Chen, Y., Guo, Y., Qiu, L.,
Liu, Z., Sun, H., Chen, S., An, Z., Zhang, Z., Li, Y., and Li, L.
(2021) Downregulation of ceramide synthase 1 promotes oral cancer
through endoplasmic reticulum stress, Int. J. Oral Sci.,
13, 10, doi: 10.1038/s41368-021-00118-4.
24.Fekry, B., Jeffries, K. A.,
Esmaeilniakooshkghazi, A., Ogretmen, B., Krupenko, S. A., and Krupenko,
N. I. (2016) CerS6 is a novel transcriptional target of p53 protein
activated by non-genotoxic stress, J. Biol. Chem., 291,
16586-16596, doi: 10.1074/jbc.M116.716902.
25.White-Gilbertson, S., Mullen, T., Senkal, C., Lu,
P., Ogretmen, B., Obeid, L., and Voelkel-Johnson, C. (2009) Ceramide
synthase 6 modulates TRAIL sensitivity and nuclear translocation of
active caspase-3 in colon cancer cells, Oncogene, 28,
1132-1141, doi: 10.1038/onc.2008.468.
26.Jensen, S. A., Calvert, A. E., Volpert, G.,
Kouri, F. M., Hurley, L. A., Luciano, J. P., Wu, Y., Chalastanis, A.,
Futerman, A. H., and Stegh, A. H. (2014) Bcl2L13 is a ceramide synthase
inhibitor in glioblastoma, Proc. Natl Acad. Sci. USA,
111, 5682-5687, doi: 10.1073/pnas.1316700111.
27.Chen, H., He, B., and Ke, F. (2022) Ceramide
synthase 6 mediates triple-negative breast cancer response to
chemotherapy through RhoA- and EGFR-mediated signaling pathways, J.
Breast Cancer, 25, 500-512, doi:
10.4048/jbc.2022.25.e47.
28.Senkal, C. E., Ponnusamy, S., Manevich, Y.,
Meyers-Needham, M., Saddoughi, S. A., Mukhopadyay, A., Dent, P.,
Bielawski, J., and Ogretmen, B. (2011) Alteration of ceramide synthase
6/C16-ceramide induces activating transcription factor 6-mediated
endoplasmic reticulum (ER) stress and apoptosis via perturbation of
cellular Ca2+ and ER/Golgi membrane network, J. Biol.
Chem., 286, 42446-42458, doi: 10.1074/jbc.M111.287383.
29.Schiffmann, S., Sandner, J., Birod, K., Wobst,
I., Angioni, C., Ruckhäberle, E., Kaufmann, M., Ackermann, H.,
Lötsch, J., Schmidt, H., Geisslinger, G., and Grösch, S.
(2009) Ceramide synthases and ceramide levels are increased in breast
cancer tissue, Carcinogenesis, 30, 745-752, doi:
10.1093/carcin/bgp061.
30.Karahatay, S., Thomas, K., Koybasi, S., Senkal,
C. E., Elojeimy, S., Liu, X., Bielawski, J., Day, T. A., Gillespie, M.
B., Sinha, D., Norris, J. S., Hannun, Y. A., and Ogretmen, B. (2007)
Clinical relevance of ceramide metabolism in the pathogenesis of human
head and neck squamous cell carcinoma (HNSCC): attenuation of
C(18)-ceramide in HNSCC tumors correlates with lymphovascular invasion
and nodal metastasis, Cancer Lett., 256, 101-111,
doi: 10.1016/j.canlet.2007.06.003.
31.Tallima, H., Azzazy, H. M. E., and El Ridi, R.
(2021) Cell surface sphingomyelin: key role in cancer initiation,
progression, and immune evasion, Lipids Health Dis., 20,
150, doi: 10.1186/s12944-021-01581-y.
32.Jing, F., Jing, C., Dai, X., Zhou, G., Di, S.,
Bi, X., Dai, T., Qin, T., and Hong, L. (2021) Sphingomyelin synthase 2
but not sphingomyelin synthase 1 is upregulated in ovarian cancer and
involved in migration, growth, and survival via different mechanisms,
Am. J. Transl. Res., 13, 4412-4421.
33.Zheng, K., Chen, Z., Feng, H., Chen, Y., Zhang,
C., Yu, J., Luo, Y., Zhao, L., Jiang, X., and Shi, F. (2019)
Sphingomyelin synthase 2 promotes an aggressive breast cancer phenotype
by disrupting the homoeostasis of ceramide and sphingomyelin, Cell
Death Dis., 10, 157, doi: 10.1038/s41419-019-1303-0.
34.Dahiya, R., Boyle, B., Goldberg, B. C., Yoon, W.
H., Konety, B., Chen, K., Yen, T. S., Blumenfeld, W., and Narayan, P.
(1992) Metastasis-associated alterations in phospholipids and fatty
acids of human prostatic adenocarcinoma cell lines, Biochem. Cell
Biol., 70, 548-554, doi: 10.1139/o92-085.
35.Li, Z., Hailemariam, T. K., Zhou, H., Li, Y.,
Duckworth, D. C., Peake, D. A., Zhang, Y., Kuo, M. S., Cao, G., and
Jiang, X. C. (2007) Inhibition of sphingomyelin synthase (SMS) affects
intracellular sphingomyelin accumulation and plasma membrane lipid
organization, Biochim. Biophys. Acta Mol. Cell Biol. Lipids,
1771, 1186-1194, doi: 10.1016/j.bbalip.2007.05.007.
36.Tafesse, F. G., Huitema, K., Hermansson, M., van
der Poel, S., van den Dikkenberg, J., Uphoff, A., Somerharju, P., and
Holthuis, J. C. (2007) Both sphingomyelin synthases SMS1 and SMS2 are
required for sphingomyelin homeostasis and growth in human HeLa cells,
J. Biol. Chem., 282, 17537-17547, doi:
10.1074/jbc.M702423200.
37.Van der Luit, A. H., Budde, M., Zerp, S., Caan,
W., Klarenbeek, J. B., Verheij, M., and Van Blitterswijk, W. J. (2007)
Resistance to alkyl-lysophospholipid-induced apoptosis due to
downregulated Sphingomyelin synthase 1 expression with consequent
Sphingomyelin- and cholesterol-deficiency in lipid rafts, Biochem.
J., 401, 541-549, doi: 10.1042/BJ20061178.
38.Itoh, M., Kitano, T., Watanabe, M., Kondo, T.,
Yabu, T., Taguchi, Y., Iwai, K., Tashima, M., Uchiyama, T., and
Okazaki, T. (2003) Possible role of ceramide as an indicator of
chemoresistance: decrease of the ceramide content via activation of
glucosylceramide synthase and Sphingomyelin synthase in chemoresistant
leukemia, Clin. Cancer Res., 9, 415-423.
39.Beier, U. H., and Görögh, T. (2005)
Implications of galactocerebrosidase and galactosylcerebroside
metabolism in cancer cells, Int. J. Cancer, 115, 6-10,
doi: 10.1002/ijc.20851.
40.Reza, S., Ugorski, M., and Suchański, J.
(2021) Glucosylceramide and galactosylceramide, small
glycosphingolipids with significant impact on health and disease,
Glycobiology, 31, 1416-1434, doi:
10.1093/glycob/cwab046.
41.García-Barros, M., Coant, N., Truman, J.
P., Snider, A. J., and Hannun, Y. A. (2014) Sphingolipids in colon
cancer, Biochim. Biophys. Acta, 1841, 773-782, doi:
10.1016/j.bbalip.2013.09.007.
42.Liu, Y. Y., Gupta, V., Patwardhan, G. A., Bhinge,
K., Zhao, Y., Bao, J., Mehendale, H., Cabot, M. C., Li, Y. T., and
Jazwinski, S. M. (2010) Glucosylceramide synthase upregulates
MDR1 expression in the regulation of cancer drug resistance
through cSrc and beta-catenin signaling, Mol. Cancer,
9, 145, 10.1186/1476-4598-9-145.
43.Joshi, P. G., and Mishra, S. (1992)
Galactocerebroside mediates Ca2+ signaling in cultured
glioma cells, Brain Res., 597, 108-113, doi:
10.1016/0006-8993(92)91511-c.
44.Pokrovskiy V. S. (2023) Human biochemistry. Lipid
turnover: a manual [in Russian], Moskva, E-noto, pp.496.
45.Lavie, Y., Cao, H., Bursten, S. L., Giuliano, A.
E., and Cabot, M. C. (1996) Accumulation of glucosylceramides in
multidrug-resistant cancer cells, J. Biol. Chem., 271,
19530-19536, doi: 10.1074/jbc.271.32.19530.
46.Lucci, A., Cho, W. I., Han, T. Y., Giuliano, A.
E., Morton, D. L., and Cabot, M. C. (1998) Glucosylceramide: a marker
for multiple-drug resistant cancers, Anticancer Res., 18,
475-480.
47.Russo, D., Parashuraman, S., and D’Angelo,
G. (2016) Glycosphingolipid-protein interaction in signal transduction,
Int. J. Mol. Sci., 17, 1732, doi:
10.3390/ijms17101732.
48.Sasaki, N., Toyoda, M., and Ishiwata, T. (2021)
Gangliosides as signaling regulators in cancer, Int. J. Mol.
Sci., 22, 5076, doi: 10.3390/ijms22105076.
49.De Maria, R., Lenti, L., Malisan, F., d'Agostino,
F., Tomassini, B., Zeuner, A., Rippo, M. R., and Testi, R. (1997)
Requirement for GD3 ganglioside in CD95- and ceramide-induced
apoptosis, Science, 277, 1652-1655, doi:
10.1126/science.277.5332.1652.
50.Veldman, R. J., Klappe, K., Hinrichs, J., Hummel,
I., van der Schaaf, G., Sietsma, H., and Kok, J. W. (2002) Altered
Sphingolipid metabolism in multidrug-resistant ovarian cancer cells is
due to uncoupling of glycolipid biosynthesis in the Golgi apparatus,
FASEB J., 16, 1111-1113, doi: 10.1096/fj.01-0863fje.
51.Xiao, S., Finkielstein, C. V., and Capelluto, D.
G. (2013) The enigmatic role of sulfatides: new insights into cellular
functions and mechanisms of protein recognition, Adv. Exp. Med.
Biol., 991, 27-40, doi: 10.1007/978-94-007-6331-9_3.
52.Suchański, J., and Ugorski, M. (2016) The
biological role of sulfatides, Postepy Hig. Med. Dosw. (Online),
70, 489-504, doi: 10.5604/17322693.1201720.
53.Su, L., Athamna, M., Wang, Y., Wang, J.,
Freudenberg, M., Yue, T., Wang, J., Moresco, E. M. Y., He, H., Zor, T.,
and Beutler, B. (2021) Sulfatides are endogenous ligands for the
TLR4-MD-2 complex, Proc. Natl. Acad. Sci. USA, 118,
e2105316118, doi: 10.1073/pnas.2105316118.
54.Takahashi, T., and Suzuki, T. (2012) Role of
sulfatide in normal and pathological cells and tissues, J. Lipid
Res., 53, 1437-1450, doi: 10.1194/jlr.R026682.
55.Boath, A., Graf, C., Lidome, E., Ullrich, T.,
Nussbaumer, P., and Bornancin, F. (2008) Regulation and traffic of
ceramide 1-phosphate produced by ceramide kinase: comparative analysis
to glucosylceramide and sphingomyelin, J. Biol. Chem.,
283, 8517-8526, doi: 10.1074/jbc.M707107200.
56.Yamaji, T., and Hanada, K. (2014) Sphingolipid
metabolism and interorganellar transport: localization of Sphingolipid
enzymes and lipid transfer proteins, Traffic, 16,
101-122, doi: 10.1111/tra.12239.
57.Baumruker, T., Bornancin, F., and Billich, A.
(2005) The role of sphingosine and ceramide kinases in inflammatory
responses, Immunol. Lett., 96, 175-185, doi:
10.1016/j.imlet.2004.09.001.
58.Bhadwal, P., Dahiya, D., Shinde, D., Vaiphei, K.,
Math, R. G. H., Randhawa, V., and Agnihotri, N. (2020) LC-HRMS based
approach to identify novel sphingolipid biomarkers in breast cancer
patients, Sci. Rep., 10, 4668, doi:
10.1038/s41598-020-61283-w.
59.Rivera, I. G., Ordoñez, M., Presa, N.,
Gangoiti, P., Gomez-Larrauri, A., Trueba, M., Fox, T., Kester, M., and
Gomez-Muñoz, A. (2016) Ceramide 1-phosphate regulates cell
migration and invasion of human pancreatic cancer cells, Biochem.
Pharmacol., 102, 107-119, doi:
10.1016/j.bcp.2015.12.009.
60.Payne, A. W., Pant, D. K., Pan, T.-C., and
Chodosh, L. A. (2014) Ceramide kinase promotes tumor cell survival and
mammary tumor recurrence, Cancer Res., 74, 6352-6363,
doi: 10.1158/0008-5472.CAN-14-1292.
61.Canals, D., Perry, D. M., Jenkins, R. W., and
Hannun, Y. A. (2011) Drug targeting of Sphingolipid metabolism:
sphingomyelinases and ceramidases, Br. J. Pharmacol.,
163, 694-712, doi: 10.1111/j.1476-5381.2011.01279.x.
62.Dyatlovitskaya, E. V., and Kandyba, A. G. (2006)
Role of biologically active sphingolipids in tumor growth,
Biochemistry (Moscow), 71, 10-17, doi:
10.1134/s0006297906010020.
63.Pulkoski-Gross, M. J., Donaldson, J. C., and
Obeid, L. M. (2015) Sphingosine-1-phoSphate metabolism: a structural
perspective, Crit. Rev. Biochem. Mol. Biol., 50, 298-313,
doi: 10.3109/10409238.2015.1039115.
64.Takabe, K., and Spiegel, S. (2014) Export of
sphingosine-1-phosphate and cancer progression, J. Lipid Res.,
55, 1839-1846, doi: 10.1194/jlr.R046656.
65.Hla, T., and Dannenberg, A. J. (2012)
Sphingolipid signaling in metabolic disorders, Cell Metab.,
16, 420-434, doi: 10.1016/j.cmet.2012.06.017.
66.Pyne, N. J., Tonelli, F., Lim, K. G., Long, J.
S., Edwards, J., and Pyne, S. (2012) Sphingosine 1-phosphate signalling
in cancer, Biochem. Soc. Transact., 40, 94-100, doi:
10.1042/bst20110602.
67.Sanchez, T., Skoura, A., Wu, M. T., Casserly, B.,
Harrington, E. O., and Hla, T. (2007) Induction of vascular
permeability by the sphingosine-1-PhoSphate receptor-2 (S1p2r) and its
downstream effectors ROCK and PTEN, Arterioscler. Thromb. Vasc.
Biol., 27, 1312-1318, doi: 10.1161/ATVBAHA.107.143735.
68.Cuvillier, O., and Levade, T. (2001) Sphingosine
1-phosphate antagonizes apoptosis of human leukemia cells by inhibiting
release of cytochrome c and Smac/Diablo from mitochondria,
Blood J. Am. Soc. Hematol., 98, 2828-2836, doi:
10.1182/blood.v98.9.2828.
69.Ponnusamy, S., Selvam, S. P., Mehrotra, S.,
Kawamori, T., Snider, A. J., Obeid, L. M., Shao, Y., Sabbadini, R., and
Ogretmen, B. (2012) Communication between host organism and cancer
cells is transduced by systemic sphingosine kinase 1/sphingosine
1-phosphate signalling to regulate tumour metastasis, EMBO Mol.
Med., 4, 761-775, doi: 10.1002/emmm.201200244.
70.Park, K. S., Kim, M. K., Lee, H. Y., Kim, S. D.,
Lee, S. Y., Kim, J. M., Ryu, S. H., and Bae, Y. S. (2007) S1p
stimulates chemotactic migration and invasion in Ovcar3 ovarian cancer
cells, Biochem. Biophys. Res. Commun., 356, 239-244, doi:
10.1016/j.bbrc.2007.02.112.
71.Dyatlovitskaya, E. V., and Kandyba, A. G. (2006)
Sphingolipids in tumor metastases and angiogenesis, Biochemistry
(Moscow), 71, 347-353, doi: 10.1134/s0006297906040018.
72.Pitson, S. M. (2011) Regulation of sphingosine
kinase and sphingolipid signaling, Trends Biochem Sci.,
36, 97-107, doi: 10.1016/j.tibs.2010.08.001.
73.Dominguez, G., Maddelein, M. L., Pucelle, M.,
Nicaise, Y., Maurage, C. A., Duyckaerts, C., Cuvillier, O., and
Delisle, M. B. (2018) Neuronal sphingosine kinase 2 subcellular
localization is altered in Alzheimer’s disease brain, Acta
Neuropathol. Commun., 6, 25, doi:
10.1186/s40478-018-0527-z.
74.Chen, H., Haddadi, N., Zhu, X., Hatoum, D., Chen,
S., Nassif, N. T., Lin, Y., and McGowan, E. M. (2022) Expression
profile of sphingosine kinase 1 isoforms in human cancer tissues and
cells: importance and clinical relevance of the neglected 1b-isoform,
J. Oncol., 7, 2250407, doi: 10.1155/2022/2250407.
75.Haddadi, N., Lin, Y., Simpson, A. M., Nassif, N.
T., and McGowan, E. M. (2017) “Dicing and Splicing”
sphingosine kinase and relevance to cancer, Int. J. Mol. Sci.,
18, 1891, doi: 10.3390/ijms18091891.
76.Diaz Escarcega, R., McCullough, L. D., and
Tsvetkov, A. S. (2021) The functional role of sphingosine kinase 2,
Front. Mol. Biosci., 8, 683767, doi:
10.3389/fmolb.2021.683767.
77.Pyne, N. J. and Pyne, S. (2010) Sphingosine 1
phosphate and cancer, Nat. Rev. Cancer, 10, 489-503, doi:
10.1038/nrc2875.
78.Kawamori, T., Kaneshiro, T., Okumura, M.,
Maalouf, S., Uflacker, A., Bielawski, J., Hannun, Y. A., and Obeid, L.
M. (2009) Role for Sphingosine kinase 1 in colon carcinogenesis,
FASEB J., 23, 405-414, doi: 10.1096/fj.08-117572.
79.Peng, J., Chen, B., Shen, Z., Deng, H., Liu, D.,
Xie, X., Gan, X., Xu, X., Huang, Z., and Chen, J. (2015) DNA promoter
hypermethylation 538 contributes to down-regulation of
galactocerebrosidase gene 539 in lung and head and neck cancers,
Int. J. Clin. Exp. Pathol., 8, 11042-11050.
80.Zhao, Y., Guo, Y., Wang, Z., Xiao, Z., Li, R.,
Luo, A., Wu, C., Jing, Z., Sun, N., Chen, X., Du, H., and Zeng, Y.
(2015) GALC gene is downregulated by promoter hypermethylation in
Epstein–Barr virus-associated nasopharyngeal carcinoma, Oncol.
Rep., 34, 1369-1378, doi: 10.3892/or.2015.4134.
81.Görögh, T., Rudert, H., Lippert, B. M.,
Gottschlich, S., Maune, S., Heidorn, K., Maass, J., Hoffmann, M.,
Meyer, J. E., Rathcke, I. O., Folz, B. J., Hortobagyi, T., and Werner,
J. A. (1999) Transcriptional repression of the human
galactocerebrosidase gene in squamous cell carcinomas of the larynx,
Int. J. Cancer, 83, 750-754, doi:
10.1002/(sici)1097-0215(19991210)83:6<750::aid-ijc9>3.0.co;2-v.
82.Sui, G., Affar, elB., Shi, Y., Brignone, C.,
Wall, N. R., Yin, P., Donohoe, M., Luke, M. P., Calvo, D., Grossman, S.
R., and Shi, Y. (2004) Yin Yang 1 (YY1) is a negative regulator of p53,
Cell, 117, 859-872, doi: 10.1016/j.cell.2004.06.004.
83.Yakovleva, T., Kolesnikova, L., Vukojević,
V., Gileva, I., Tan-No, K., Austen, M., Lüscher, B., Ekström,
T. J., Terenius, L., and Bakalkin, G. (2004) YY1 binding to a subset of
p53 DNA-target sites regulates p53-dependent transcription, Biochem.
Biophys. Res. Commun., 318, 615-624, doi:
10.1016/j.bbrc.2004.04.065.
84.Presta, M. (2021) β-Galactosylceramidase in
cancer: friend or foe? Trends Cancer, 7, 974-977, doi:
10.1016/j.trecan.2021.08.001.
85.Belleri, M., Paganini, G., Coltrini, D., Ronca,
R., Zizioli, D., Corsini, M., Barbieri, A., Grillo, E., Calza, S.,
Bresciani, R., Maiorano, E., Mastropasqua, M. G., Annese, T.,
Giacomini, A., Ribatti, D., Casas, J., Levade, T., Fabrias, G., and
Presta, M. (2020) β-Galactosylceramidase promotes melanoma growth
via modulation of ceramide metabolism, Cancer Res., 80,
5011-5023, doi: 10.1158/0008-5472.CAN-19-3382.
86.Barth, B. M., Shanmugavelandy, S. S., Tacelosky,
D. M., Kester, M., Morad, S. A., and Cabot, M. C. (2013)
Gaucher’s disease and cancer: a sphingolipid perspective,
Crit. Rev. Oncog., 18, 221-234, doi:
10.1615/critrevoncog.2013005814.
87.Henry, B., Ziobro, R., Becker, K. A., Kolesnick,
R., and Gulbins, E. (2013) Acid sphingomyelinase, Handb. Exp.
Pharmacol., 215, 77-88, doi:
10.1007/978-3-7091-1368-4_4.
88.Hertervig, E., Nilsson, A., Nyberg, L., and Duan,
R. D. (1997) Alkaline sphingomyelinase activity is decreased in human
colorectal carcinoma, Cancer, 79, 448-453, doi:
10.1002/(sici)1097-0142(19970201)79:3<448::aid-cncr4>3.0.co;2-e.
89.Demircan, B., Dyer, L. M., Gerace, M.,
Lobenhofer, E. K., Robertson, K. D., and Brown, K. D. (2009)
Comparative epigenomics of human and mouse mammary tumors, Genes
Chromosomes Cancer, 48, 83-97, doi: 10.1002/gcc.20620.
90.Clarke, C. J. (2018) Neutral sphingomyelinases in
cancer, Sphingolipids Cancer, 140, 97-119, doi:
10.1016/bs.acr.2018.04.010.
91.Revill, K., Wang, T., Lachenmayer, A., Kojima,
K., Harrington, A., Li, J., Hoshida, Y., Llovet, J. M., and Powers, S.
(2013) Powers genome-wide methylation analysis and epigenetic unmasking
identify tumor suppressor genes in hepatocellular carcinoma,
Gastroenterology, 145, 1424-1435, doi:
10.1053/j.gastro.2013.08.055.
92.Kim, W. J., Okimoto, R. A., Purton, L. E.,
Goodwin, M., Haserlat, S. M., Dayyani, F., Sweetser, D. A., McClatchey,
A. I., Bernard, O. A., Look, A. T., Bell, D. W., Scadden, D. T., and
Haber, D. A. (2008) Mutations in the neutral sphingomyelinase gene
SMPD3 implicate the ceramide pathway in human leukemias, Blood,
111, 4716-4722, doi: 10.1182/blood-2007-10-113068.
93.Marchesini, N., and Hannun, Y. A. (2004) Acid and
neutral sphingomyelinases: roles and mechanisms of regulation,
Biochem. Cell Biol., 82, 27-44, doi: 10.1139/o03-091.
94.Megha, and London, E. (2004) Ceramide selectively
displaces cholesterol from ordered lipid domains (rafts): implications
for lipid raft structure and function, J. Biol. Chem.,
279, 9997-10004, doi: 10.1074/jbc.M309992200.
95.Hannun, Y. A., and Obeid, L. M. (2018)
Sphingolipids and their metabolism in physiology and disease, Nat.
Rev. Mol. Cell Biol., 19, 175-191, doi:
10.1038/nrm.2017.107.
96.Jarvis, W. D., Kolesnick, R. N., Fornari, F. A.,
Traylor, R. S., Gewirtz, D. A., and Grant, S. (1994) Induction of
apoptotic DNA damage and cell death by activation of the Sphingomyelin
pathway, Proc. Natl. Acad. Sci. USA, 91, 73-77, doi:
10.1073/pnas.91.1.73.
97.Aslan, M., Afsar, E., Kirimlioglu, E., Ceker, T.,
and Yilmaz, C. (2021) Antiproliferative effects of thymoquinone in
MCF-7 breast and HepG2 liver cancer cells: possible role of ceramide
and ER stress, Nutr. Cancer, 73, 460-472, doi:
10.1080/01635581.2020.1751216.
98.Kilkus, J., Goswami, R., Testai, F. D., and
Dawson, G. (2003) Ceramide in rafts (detergent-insoluble fraction)
mediates cell death in neurotumor cell lines, J. Neurosci. Res.,
72, 65-75, doi: 10.1002/jnr.10549.
99.Rotolo, J. A., Zhang, J., Donepudi, M., Lee, H.,
Fuks, Z., and Kolesnick, R. (2005) Caspase-dependent and -independent
activation of acid sphingomyelinase signaling, J. Biol. Chem.,
280, 26425-26434, doi: 10.1074/jbc.M414569200.
100.Ferranti, C. S., Cheng, J., Thompson, C.,
Zhang, J., Rotolo, J. A., Buddaseth, S., Fuks, Z., and Kolesnick, R. N.
(2020) Fusion of lysosomes to plasma membrane initiates
radiation-induced apoptosis, J. Cell Biol., 219,
e201903176, doi: 10.1083/jcb.201903176.
101.Clarke, C. J., Shamseddine, A. A., Jacob, J.
J., Khalife, G., Burns, T. A., and Hannun, Y. A. (2016) ATRA
transcriptionally induces nSMase2 through CBP/p300-mediated histone
acetylation, J. Lipid Res., 57, 868-881, doi:
10.1194/jlr.m067447.
102.Morad, S. A. and Cabot, M. C. (2013)
Ceramide-orchestrated signalling in cancer cells, Nat. Rev.
Cancer, 13, 51-65, doi: 10.1038/nrc3398.
103.Tan, S. F., Liu, X., Fox, T. E., Barth, B. M.,
Sharma, A., Turner, S. D., Awwad, A., Dewey, A., Doi, K., Spitzer, B.,
Shah, M. V., Morad, S. A., Desai, D., Amin, S., Zhu, J., Liao, J., Yun,
J., Kester, M., Claxton, D. F., Wang, H. G., and Loughran, T. P., Jr.
(2016) Acid ceramidase is upregulated in AML and represents a novel
therapeutic target, Oncotarget, 7, 83208-83222, doi:
10.18632/oncotarget.13079.
104.Xu, R., Jin, J., Hu, W., Sun, W., Bielawski,
J., Szulc, Z., Taha, T., Obeid, L. M., and Mao, C. (2006) Golgi
alkaline ceramidase regulates cell proliferation and survival by
controlling levels of sphingosine and S1P, FASEB J., 20,
1813-1825, doi: 10.1096/fj.05-5689com.
105.Coant, N., Sakamoto, W., Mao, C., and Hannun,
Y. A. (2017) Ceramidases, roles in sphingolipid metabolism and in
health and disease, Adv. Biol. Regul., 63, 122-131, doi:
10.1016/j.jbior.2016.10.002.
106.Maeda, I., Takano, T., Matsuzuka, F., Maruyama,
T., Higashiyama, T., Liu, G., Kuma, K., and Amino, N. (1999) Rapid
screening of specific changes in mRNA in thyroid carcinomas by sequence
specific-differential display: decreased expression of acid ceramidase
mRNA in malignant and benign thyroid tumors, Int. J. Cancer,
81, 700-704, doi:
10.1002/(sici)1097-0215(19990531)81:5<700::aid-ijc5>3.0.co;2-d.
107.Roh, J. L., Park, J. Y., Kim, E. H., and Jang,
H. J. (2016) Targeting acid ceramidase sensitises head and neck cancer
to cisplatin, Eur. J. Cancer, 52, 163-172, doi:
10.1016/j.ejca.2015.10.056.
108.Poterala-Hejmo, A., Golda, A., Pacholczyk, M.,
Student, S., Tylki-Szymańska, A., and Lalik, A. (2020) A
possible role for arylsulfatase G in dermatan sulfate metabolism,
Int. J. Mol. Sci., 21, 4913, doi:
10.3390/ijms21144913.
109.Kolter, T., and Sandhoff, K. (2005) Principles
of lysosomal membrane digestion: stimulation of Sphingolipid
degradation by sphingolipid activator proteins and anionic lysosomal
lipids, Annu. Rev. Cell Dev. Biol., 21, 81-103, doi:
10.1146/annurev.cellbio.21.122303.120013.
110.Garcia, J., Callewaert, N., and Borsig, L.
(2007) P-selectin mediates metastatic progression through binding to
sulfatides on tumor cells, Glycobiology, 17, 185-196,
doi: 10.1093/glycob/cwl059.
111.Simonis, D., Schlesinger, M., Seelandt, C.,
Borsig, L., and Bendas, G. (2010) Analysis of SM4 sulfatide as a
P-selectin ligand using model membranes, Biophys. Chem.,
150, 98-104, doi: 10.1016/j.bpc.2010.01.007.
112.Waggoner, D. W., Gómez-Muñoz, A.,
Dewald, J., and Brindley, D. N. (1996) Phosphatidate phosphohydrolase
catalyzes the hydrolysis of ceramide 1-phosphate, lysophosphatidate,
and sphingosine 1-phosphate, J. Biol. Chem., 271,
16506-16509, doi: 10.1074/jbc.271.28.16506.
113.Boudker, O., and Futerman, A. H. (1993)
Detection and characterization of ceramide-1-phoSphate phosphatase
activity in rat liver plasma membrane, J. Biol. Chem.,
268, 22150-22155.
114.Long, J., Darroch, P., Wan, K. F., Kong, K. C.,
Ktistakis, N., Pyne, N. J., and Pyne, S. (2005) Regulation of cell
survival by lipid phosphate phosphatases involves the modulation of
intracellular phosphatidic acid and sphingosine 1-phosphate pools,
Biochem. J., 391, 25-32, doi: 10.1042/BJ20050342.
115.Zhao, Y., Usatyuk, P. V., and Cummings, R.
(2005) Lipid phosphate phosphatase-1 regulates lysophosphatidic
acid-induced calcium release, NF-kappaB activation and interleukin-8
secretion in human bronchial epithelial cells, Biochem. J.,
385, 493-502, doi: 10.1042/BJ20041160.
116.Kihara, A., Mitsutake, S., Mizutani, Y., and
Igarashi, Y. (2007) Metabolism and biological functions of two
phosphorylated sphingolipids, sphingosine 1-phosphate and ceramide
1-phosphate, Prog. Lipid Res., 46, 126-144, doi:
10.1016/j.plipres.2007.03.001.
117.Uranbileg, B., Kurano, M., Kano, K., Sakai, E.,
Arita, J., Hasegawa, K., Nishikawa, T., Ishihara, S., Yamashita, H.,
Seto, Y., Ikeda, H., Aoki, J., and Yatomi, Y. (2022) Sphingosine
1-phosphate lyase facilitates cancer progression through converting
sphingolipids to glycerophospholipids, Clin. Transl. Med.,
12, e1056, doi: 10.1002/ctm2.1056.
118.Hibbs, K., Skubitz, K. M., Pambuccian, S. E.,
Casey, R. C., Burleson, K. M., Oegema, T. R., Jr., Thiele, J. J.,
Grindle, S. M., Bliss, R. L., and Skubitz, A. P. (2004) Differential
gene expression in ovarian carcinoma: identification of potential
biomarkers, Am. J. Pathol., 165, 397-414, doi:
10.1016/S0002-9440(10)63306-8.
119.Uranbileg, B., Ikeda, H., Kurano, M., Enooku,
K., Sato, M., Saigusa, D., Aoki, J., Ishizawa, T., Hasegawa, K.,
Kokudo, N., and Yatomi, Y. (2016) Increased mRNA levels of sphingosine
kinases and S1P lyase and reduced levels of S1P were observed in
hepatocellular carcinoma in association with poorer differentiation and
earlier recurrence, PloS One, 11, e0149462, doi:
10.1371/journal.pone.0149462.
120.Uranbileg, B., Nishikawa, T., Ikeda, H.,
Kurano, M., Sato, M., Saigusa, D., Aoki, J., Watanabe, T., and Yatomi,
Y. (2018) Evidence suggests sphingosine 1-phosphate might be actively
generated, degraded, and transported to extracellular spaces with
increased S1P2 and S1P3 expression in colon cancer, Clin. Colorectal
Cancer, 17, e171-e182, doi: 10.1016/j.clcc.2017.11.004.
121.Oskouian, B., Sooriyakumaran, P., Borowsky, A.
D., Crans, A., Dillard-Telm, L., Tam, Y. Y., Bandhuvula, P., and Saba,
J. D. (2006) Sphingosine-1-phosphate lyase potentiates apoptosis via
p53‐ and p38‐dependent pathways and is down-regulated in
colon cancer, Proc. Natl. Acad. Sci. USA, 103,
17384-17389, doi: 10.1073/pnas.0600050103.