Plant Telomeres and Telomerases. A Review
T. D. McKnight,1,2 M. S. Fitzgerald,3 and D. E. Shippen3
1Department of Biology, Texas A&M University, College Station, Texas 77843, USA; fax: 409-845-3896; E-mail: mcknight@bio.tamu.edu2To whom correspondence should be addressed.
3Department of Biochemistry and Biophysics, Texas A&M University, College Station, Texas 77843, USA.
Submitted July 25, 1997.
Barbara McClintock began investigating plant telomeres during the 1930s, but little additional work was done in this area until a telomeric DNA sequence was isolated and characterized from Arabidopsis thaliana in 1988. This sequence, a simple repeat of the heptanucleotide 5´-TTTAGGG-3´, has been found in telomeres of almost all plants analyzed. Telomere length in plants, which can be a long as 75 kb or as short as 2 kb, is controlled by both genetic and developmental factors. The major mechanism for synthesis of telomeres is telomerase, a ribonucleoprotein with reverse transcriptase activity. Telomerase expression is highly regulated in both plants and animals. For example, there is little or no detectable expression of telomerase in most vegetative tissues of plants nor in most somatic tissues of animals. In contrast to animals, plants do not specify a germ line until late in development, but telomerase is reactivated during flowering, possibly to ensure that gametes and embryos arising from them inherit fully functional chromosomes. Telomerase is also highly expressed in plant tissue culture cells, as might be expected for cells with an unlimited capacity for proliferation. Despite recent progress in investigating plant telomeres and telomerase at the molecular level, there is still much more to learn, especially concerning the developmental control of telomerase activity.
KEY WORDS: plants, telomerase, telomere, chromosomes, plant development.
Abbreviations: RFLP) restriction fragment length polymorphism; ISH) in situ hybridization; TRAP) telomere repeat amplification protocol; PCR) polymerase chain reaction.
Telomeres are nucleoprotein caps at the ends of linear eukaryotic
chromosomes that are essential for maintaining the integrity of the
genome. Because the ends of linear chromosomes cannot be replicated by
DNA-dependent DNA polymerases, telomeric DNA must be generated and
maintained by a different mechanism. The primary mechanism for telomere
DNA synthesis occurs through the action of telomerase, a
ribonucleoprotein with reverse transcriptase activity [1]. Telomerase adds telomeric DNA onto the 3´
ends of chromosomes by copying a short template sequence within its RNA
subunit. Telomerase is developmentally regulated in mammals, and high
expression levels are associated with tissues that have the capacity
for extensive cellular replication and regeneration. In most somatic
cells telomerase activity is undetectable or very low [2].
Telomere structure in most plants is very similar to that of mammals. Plants, however, have fundamentally different patterns of development from animals, and this difference may be reflected in the way telomere synthesis by telomerase is regulated. Plants have a highly plastic developmental pattern, most notably characterized by the fact that plants do not specify a germ line until very late. This delayed designation of reproductive cells is probably an adaptation to the sessile nature of plants and the fact that they cannot escape herbivores or physical agents that could damage reproductive structures that had to be maintained throughout the life of the plant. Damage to the apical meristem, which in many plants eventually generates flowers, stimulates previously quiescent meristems to assume the growth and reproductive functions of the damaged meristem. Because these axillary meristems may be pressed into service at anytime, they must be capable of maintaining or generating functional telomeres.
Telomere shortening, presumably through attrition caused by the inability of the usual DNA replication machinery to replicate the very ends of linear chromosomes, occurs throughout the life of mammals and is associated with aging [3]. Some plants have much longer life spans than animals, and therefore must maintain intact telomeres for much longer periods. Bristle-cone pines (Pinus aristata), for example, can live for several thousand years, and one living tree was found to be at least 4844 years old [4]. Alerce trees (Fitzroya cupressoides) also have life spans measured in millennia [5].
Plant cells are totipotent. Even highly differentiated cells such as guard cells can undergo dedifferentiation and redifferentiation to give rise to whole plants [6]. Again, these cells must either maintain or generate a full complement of telomeric repeats. Totipotency has been considered to be a distinct difference between plants and animals, but with the recent cloning of mammals [7], it may not be such a great difference after all. It will be interesting, however, to compare the length of telomeres between the younger and older mammalian clones.
Plant telomeres were investigated initially by Barbara McClintock at about the same time that Hermann Muller was examining Drosophila telomeres [8, 9]. The work of these two pioneers demonstrated that telomeres served to cap and protect the ends of chromosomes from degradation and also to prevent fusion of chromosomes ends.
Using genetic and cytogenetic approaches, McClintock found that dicentric chromosomes broken during anaphase in the endosperm of developing maize kernels could rejoin, or fuse, through their newly formed ends to recreate another dicentric chromosome. Such chromosomes would undergo repeated rounds of breakage and fusion (the classic "breakage--fusion--bridge" cycle) with the eventual loss of telomeric genes from some cell lineages. The loss of telomere proximal genes was detected by the unmasking of recessive genes on a normal, monocentric chromosome which gave rise to visible sectors. The triploid endosperm tissue, which makes up the bulk of the maize kernel, produced many sectors, as would be expected if genes were sequentially lost through repeated rounds of breakage and fusion. In contrast, the embryo (or the plant developed from it) did not contain any sectored tissues, suggesting a uniformity of chromosomes that could occur only if the breakage--fusion--bridge cycle ceased very early in embryonic development. Cytological analysis confirmed that cells in all tissues of the plant received identical chromosomes, and that none of these were dicentric. McClintock concluded that broken chromosome ends in the embryo, but not the endosperm, were healed and converted to normal chromosome ends that did not fuse. In the endosperm, where healing does not occur, the breakage--fusion--bridge cycle continues indefinitely [8, 9].
McClintock had no real explanation for the different behavior of broken chromosome ends in endosperm and embryo tissue, but we can now speculate that telomerase is active in the embryo where it heals newly produced ends and is absent in the endosperm. Recent studies in other monocots confirm that broken chromosomes are healed by the addition of telomeres to the newly formed end, but this de novo telomere synthesis is restricted to gametogenesis or early stages of embryogenesis [10].
Endosperm may not require telomerase activity because of its ephemeral nature. If, however, telomerase is not active in endosperm, then telomeres in this tissue are predicted to be shorter than those in the embryo. This difference would be exacerbated by the endoreduplication that increases the ploidy level of the endosperm from three to near one hundred [11], but so far no one has compared telomere lengths between embryo and endosperm.
Telomere Structure in Plants
Few studies on plant telomeres were published after McClintock's pioneering observations until an Arabidopsis thaliana telomere was cloned by Richards and Ausubel in 1988 [12]. After using an innovative cloning procedure to make a genomic library enriched for telomeric sequences, they searched for clones that hybridized to Bal-31 exonuclease-sensitive regions of the genome. The single clone isolated by this procedure, pAtT4, contains 53 copies of the heptanucleotide 5´-TTTAGGG-3´ and two copies of the closely related variant 5´-TTTAGAG-3´. The telomeric nature of this clone was confirmed by using it to detect an RFLP4 (restriction fragment length polymorphism) in a telotrisomic line and in a population segregating for the extra telocentric chromosome [12].
The Arabidopsis telomere was the first one characterized in any multicellular eukaryote, and Richards and Ausubel used it to determine how widespread this sequence was. They found that it hybridized to exonuclease-sensitive regions in the maize and human genomes, but it did not hybridize to anything at all in the Drosophila genome [12]. Subsequently, human telomeres were isolated and found to contain a similar hexanucleotide repeat TTAGGG [13], and we now know that Drosophila telomeres are atypical and are composed of two retroposons [14].
Richards et al. [15] used a second cloning method, complementation of a telomere-deficient yeast artificial chromosome, to isolate additional Arabidopsis telomere clones. Two new clones were isolated, and both showed large-scale structure that was not apparent in their previously described telomere clone. The YpAtT1 clone ended with a few copies of the yeast telomeric repeat that were added by the host. Just interior to these yeast sequences are 17 identical copies of the canonical TTTAGGG repeat, which are followed by about 40 copies that differ by only one nucleotide (TTAAGGG is the most common variant). Moving further toward the centromere, there are about 10 relatively degenerate versions of the canonical telomere repeat. There is a clear boundary between these telomeric sequences and the adjacent purine-rich telomere-associated sequence [15].
The second telomeric clone, YpAtT7, also ended with yeast telomere repeats, but the adjacent Arabidopsis sequence was composed of 26 variant repeats (mostly TTCAGGG). This variant region was then flanked on the centromeric side by more than 40 copies of the typical TTTAGGG repeat. As was the case for the first clone, this stretch of perfect repeats was followed by a block of variant repeats (often TTCAGGG) and then a block of more degenerate repeats before the boundary with the telomere-associated sequence [15].
Subsequent characterization of telomeres from maize [16], tomato [17], and the unicellular green alga Chlorella vulgaris [18] demonstrated that they too are composed of TTTAGGG repeats. In situ hybridization (ISH) has been used to show that this type of telomere is widespread throughout the plant kingdom. Probably the most comprehensive study is that reported by Fuchs et al. [19], who looked at 44 species from 14 families covering angiosperms, gymnosperms, and bryophytes. In all but one family, a (TTTAGGG)n probe hybridized to the ends of chromosomes indicating that Arabidopsis-type telomeres are nearly universal. Even telomeres of Chlamydomonas reinhardtii, an alga only distantly related to land plants, are composed of the related octanucleotide TTTTAGGG [20].
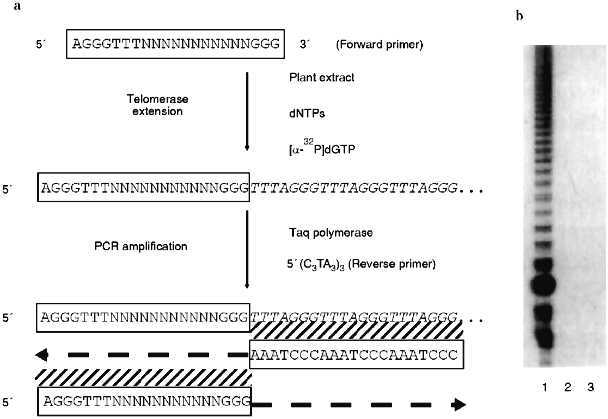
In their survey of 14 plant families, Fuchs et al. [19] examined five species in the onion family (Alliaceae) and found that the Arabidopsis telomere probe did not hybridize to any chromosomes from these plants. Southern blots presented in a later paper [21] confirmed a complete lack of Arabidopsis-type telomeric sequences in the onion genomes. Furthermore, telomerase assays using the telomeric repeat amplification protocol (TRAP; see Fig. 1) with callus from garlic (Allium sativum) failed to detect any enzyme activity (J. Burroughs, T. D. McKnight, D. E. Shippen, unpublished results). A negative result with the TRAP assay, which is probably more sensitive than ISH, could be due to the presence of a different telomeric repeat in garlic that is not recognized by the reverse primer for the PCR (polymerase chain reaction) step of the TRAP assay (Fig. 1), or it may reflect that fact that there is no telomerase activity present in the suspension cells.
Taken together, the data argue that telomeres in plants in the onion family may not be generated by a typical telomerase enzyme. A non-telomerase mechanism for maintaining telomeres has been well documented in Drosophila. In this case, the retroposons HeT-A and TART comprise the bulk of the chromosome termini [14]. Pich et al. [21] used ISH to localize a 375-bp satellite to the ends of most chromosomes in two onion species. The only chromosome ends that did not hybridize to the satellite repeats did hybridize to a ribosomal DNA probe. The repetitive nature of the satellite and rDNA loci would facilitate recombination-based mechanisms that could prevent shortening of the chromosomes during replication. Given the limits of resolution of ISH on condensed chromosomes, however, it is unclear whether the satellite and rDNA loci constitute the actual ends of the chromosomes or whether some as yet undetected sequence functions as the telomere. In Arabidopsis, which has prototypical telomeres, both rDNA clusters have been mapped by RFLPs to the extreme termini of chromosomes, and single restriction fragments of 8 and 13 kb hybridize to probes for both telomere repeats and rDNA. For one of these telomeres, DNA sequencing showed that the rDNA continued right up to the start of the telomeric repeats [22]. Whether Alliaceae chromosomes actually terminate in rDNA loci, or whether other sequences are present beyond them, it is clear that plants, like animals, can have atypical telomeres.Fig. 1. Telomere repeat amplification protocol (TRAP). a) A schematic illustration of the TRAP assay. A forward primer is mixed with a partially purified extract of plant nuclei, dNTPs. The forward primer is recognized by telomerase, and new telomeric repeats are synthesized onto the 3´ end by the enzyme’s reverse transcriptase activity. A reverse primer complementary to the plant telomeric sequence is then added to the reaction cocktail. PCR is performed to increase the sensitivity of the assay. Reaction products are resolved by fractionation on an acrylamide gel and visualized by autoradiography. b) A representative TRAP assay. The TRAP assay was performed with 500 ng of protein from Arabidopsis thaliana suspension culture (lane 1), 500 ng of protein from the leaves of Nicotiana tabacum (lane 2), and without plant extract (lane 3). The telomerase mediated synthesis of a seven-nucleotide ladder of products is detected only with the Arabidopsis culture.
In most plants where it has been examined, repetitive sequences elements are found interior to the telomeric repeats [17, 23-25]. This is not surprising, since most plant genomes are composed largely of repeated elements. These subtelomeric repeats may have no functional significance. In yeast, two distinct repetitive elements are associated with telomeres, but strains in which these subterminal repeats have been eliminated grow normally (unpublished results cited in [26]). Similarly, plant telomeres do not require special subterminal sequences, as shown by the Arabidopsis telomere where rDNA sequences are immediately adjacent to the TTTAGGG repeats [22].
Proteins that bind to telomeric DNA are also required for the proper structure and function of telomeres, but very little is known about the protein component of plant telomeres. Zentgraf [27] isolated proteins from Arabidopsis nuclei that bind to plant telomeric repeats by running electrophoretic mobility shift assays and recovering the DNA--protein complexes. The proteins from these shifted bands were characterized by additional rounds of electrophoresis and found to consist of a 67 kD band that contained proteins with three distinct, but very close isoelectric points. It has not been determined whether these proteins are slight modifications of a single gene product or whether they are derived from different genes. The Arabidopsis protein does not appear to be a component of telomerase because treatment of nuclear extracts with RNase did not affect binding to the telomeric repeats [27]. The relation of the Arabidopsis protein to previously characterized telomere binding proteins such as TRF1 in mammals [28], RAP1 in yeast (reviewed in [29]), or the telomeric proteins of hypotrichous ciliates [30] is not clear. However, a protein sequence element called the "telobox", which is a hallmark of the mammalian TRF, is found in proteins encoded by expressed sequence tags (ESTs, Expressed Sequence Tags) in Arabidopsis and rice [31].
Regulation of Telomere Length in Plants
Although the telomere repeat is conserved throughout land plants, the actual length of the telomeres is highly variable. At least some plant telomeres shorten during development, just as mammalian telomeres do. Kilian et al. [32] reported that barley telomeres shorten even during embryo development. The youngest embryos assayed (less than 1 mm long) had telomeres approximately 75 kb long. As the embryos grew, their telomeres decreased to about 25 kb. After germination, the leaves had telomeres just slightly shorter than the oldest embryos (20 kb). Dot blots were used to confirm that this shortening of terminal restriction fragments was due to a loss of telomeric repeats and not to changes in other sequences on the terminal restriction fragments.
Telomere length was also measured in the barley inflorescence spike, where it decreased from 40 to 45 kb in young spikes to about 20 kb in older spikes. Because telomeres in young embryos are much longer than they are in the mature inflorescence spike, dramatic lengthening must occur in or before early embryogenesis, possibly, during flowering and gametogenesis. The high levels of telomerase activity detected in cauliflower floral buds [33] are consistent with the reactivation of telomerase upon flowering.
The 50 kb decrease in telomere length during barley embryogenesis and the 20 kb decrease during spike growth probably reflect a developmentally programmed decrease, since shortening of telomeres through attrition in the absence of telomerase could not account for this much change. Neither the mechanism nor the rationale for such a large decrease is known, but extrachromosomal elements containing telomeric sequences have been observed in mature wheat embryos [34], implying that large portions of telomeric DNA can be cleaved from the chromosomes. These elements, although stable in mature dormant embryos, are eliminated within the first three hours of germination [34]. If such extrachromosomal elements are produced during embryogenesis, it may be possible to detect them by ISH.
In contrast to the shortening of telomeres during growth of differentiated barley tissue, telomeres grow to 300 kb during undifferentiated growth in suspension culture [32]. As might be predicted for cells with an extensive capacity for proliferation, telomerase activity is very high in suspension cultures of many plants [33]. However, high levels of telomerase do not necessarily culminate in excessively long telomeres. In a rapidly dividing tobacco cell culture that had been maintained for 24 years, telomeres ranged from 20 to 130 kb, about the same range as in tobacco leaves (40-160 kb). In contrast, most of the telomeres from cultured cells were in the 20- to 50-kb region, while those from leaves were in the 90- to 130-kb region. Furthermore, although telomerase activity was detected in the cultured cells, telomere length was stable over a six-month period that covered approximately 225 doublings [35].
Broun et al. [36] reported that telomeres in tomato leaves do not change in size over a five-month period beginning when the plants are one month old. This apparent stability, similar to that of tobacco cell cultures but different from the shortening seen in barley development [32], could be a fundamental difference between monocots and dicots, or it may be an adaptation to the fact that most tomato varieties have an indeterminate growth habit and can continue to grow almost indefinitely.
In addition to developmental control of telomere length, there also appears to be significant genetic control of telomere length in plants. Burr et al. [16] found a 25-fold difference in the lengths of telomeres among inbred lines of maize, ranging from less than 2 kb for the WF9 line to about 40 kb for the CM37 line. Inheritance of telomere length was investigated by examining a recombinant inbred (RI) family derived from parents with long (about 13 kb) or short (about 7.5 kb) telomeres. The F1 progeny from this cross have short (8.1 kb) telomeres, but individual RI lines have telomeres as short as 4.5 kb and as long as 20 kb, extending well beyond the range of the parental lines. Telomere lengths were stable within each RI line when measured over the seventh and eighth generations and appeared to be inherited in a complex quantitative manner. Three genetic loci could account for half of the variation [16]. Additional loci that have a minor effect must be involved, but their detection will require analysis of larger populations.
Plant Telomerases
In most eukaryotes, with Drosophila and possibly Allium as notable exceptions, telomeres are synthesized and maintained by telomerase. Although telomerase has not yet been purified from any multicellular eukaryote, several telomere-associated proteins, including one that appears to contain the reverse transcriptase active site [37], have been reported [38]. The telomerase RNA subunit has been well characterized in a number of systems. Telomerase RNAs, which range from 150 nucleotides in ciliates [39, 40] to approximately 1400 nucleotides in yeast [41], harbor a short domain that is complementary to about one and a half copies of the telomere repeat. This region of the RNA forms Watson--Crick base pairs with the 3´ terminus of the chromosome and provides a template for the polymerization of telomeric repeats.
The identification of telomerase in plants was reported by three independent groups in 1996. Fajkus et al. [35] used the conventional biochemical telomerase assay method that is based on extension of a telomere-like primer to detect activity in a tobacco cell line. Telomerase extends the primers in multiples of the telomere repeat. When the products of the reaction are resolved on a DNA sequencing gel they produce a ladder of bands reflecting rearrangements of the enzyme and primer as each repeat is synthesized. Hence, the periodicity of the ladder typically corresponds to the length of the telomere repeat. Fajkus et al. [35] observed broad smears of extended products that were longer than the initial primer by approximately one, four, and six telomeric repeats. These products probably were produced by telomerase (or at least some type of reverse transcriptase) because they were not produced if the extract was treated with RNase before the assay.
The recently described TRAP [42] seems to be the best way to detect telomerase activity in plants and has been used to assay telomerase in a variety of plant tissues. In the TRAP assay, telomerase extends a primer with little homology to the telomeric repeat and the products of this reaction are amplified by PCR. The reverse primer for PCR consists of several copies of the telomeric repeat expected for the organism. The TRAP assay is outlined in Fig. 1.
Heller et al. [43] used the TRAP assay to show that telomerase is present in barley embryos, anthers, and carpels, but not detectable in barley leaves. Telomerase was also detected in immature seeds of Arabidopsis. In all cases, products of the TRAP assay were not produced if the plant extracts were treated with RNase.
Our laboratories also used TRAP assays to characterize plant telomerase activity and survey its expression in various tissues. Several plant telomerases, especially telomerase from cauliflower inflorescences, have a relatively relaxed primer specificity. In contrast to ciliate and mammalian telomerases, these plant enzymes will add telomeric repeats onto primers bearing no 3´ complementarity to the telomerase RNA template. This ability to initiate telomere synthesis in the absence of Watson--Crick base pairing between the primer and the telomerase RNA template may account for de novo telomere formation observed with broken chromosomes in maize [8, 9], wheat [10], and barley [44].
Because plant telomerase activity (as analyzed by the TRAP assay) is very similar to that found in most other eukaryotes, the structure of plant telomerase should be very similar also. Telomerase has not been purified from any plant source yet, but the abundant activity in suspension cultures and in cauliflower heads makes them the best sources of the enzyme. Unlike animal cell cultures, plant cell cultures can easily be scaled up to provide several kilograms of starting material, and cauliflower heads are commercially available.
Telomerase Expression Patterns in Plants
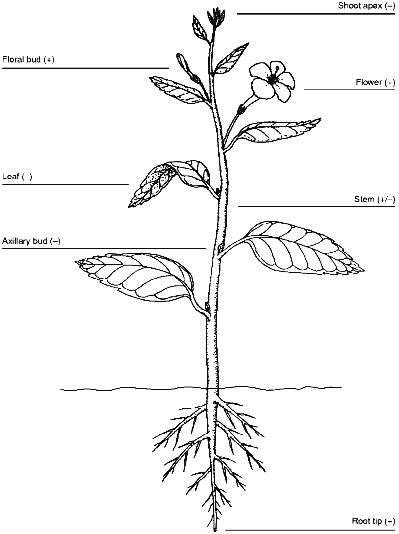
If telomerase is developmentally controlled in plants as it is in mammals, activity should be present in plant tissues with long-term proliferative potential, such as the root and shoot meristems and the vascular cambium, but absent from mature tissues such as leaves. For the most part, results with soybean plants support this conclusion [33]. Telomerase activity was detected in root tips of both 3-day-old seedlings and older plants, and there was low, but detectable, activity in stems, probably associated with the vascular cambium. As expected, there was no detectable telomerase activity in either the primary or trifoliate leaves. Surprisingly, there was no telomerase activity in either the shoot apex or axillary buds. Both of these regions contain meristematic cells, and although those in the axillary buds are quiescent, the shoot meristem is a major site of cell division. Mixing experiments showed that the shoot apex did not contain inhibitors of either telomerase or Taq DNA polymerase used in the PCR step of the TRAP assay [33]. It is possible that telomerase is expressed in the apical meristem, but in too few cells to be detected. If telomerase is not expressed in the meristem, then telomeres should be shorter in leaves at the top of the plant than at the bottom of the plant. This possibility has not been tested yet in soybeans, but the shortening of telomeres in barley inflorescence spikes [32] and the stability of telomere length in tomato leaves [36] suggests that telomerase may be active in the meristems of some plants and not others.
Chromosome healing in maize embryos and early wheat development suggests that telomerase should be active during reproduction [8, 10]. Recent studies confirm this. Heller et al. [43] detected telomerase in barley embryos and in both male and female floral organs. They did not examine inflorescence spikes for telomerase activity, but because telomeres shorten during growth of the barley inflorescence spike, telomerase may not be turned on until flowering. Telomerase activity may be restricted to early embryogenesis or gametogenesis, because telomeres shorten during development of barley embryos [32]. Fitzgerald et al. [33] found copious telomerase activity in cauliflower floral buds, somewhat less activity in the subtending peduncle, and very little activity in the stems and leaves, consistent with the idea that telomerase is repressed in vegetative tissues but reactivated during reproduction. The expression pattern of telomerase in plants is summarized in Fig. 2.
As predicted from the results of Kilian et al. [32], who reported excessively long telomeres in barley suspension cultures, high levels of telomerase activity are present in suspension cultures of carrot, soybean, Arabidopsis and rice [33]. Plant cells grown under these conditions are immortal, rapidly dividing, and relatively dedifferentiated so the high amount of telomerase in them can be compared to that seen in most mammalian cancer cells [42].Fig. 2. Patterns of telomerase expression in plants. Expression of telomerase in various plant organs is indicated by plus or minus signs. Weak telomerase activity, probably associated with the vascular cambium, is indicated for the stem. Data are summarized from [33] and [43].
Future Directions
Despite recent progress in investigating plant telomeres and telomerase at the molecular level, there is still much more to learn, especially concerning the developmental control of telomerase. While little or no telomerase activity is present in somatic tissues, including shoot apices, telomerase is apparently activated during flowering. Heller et al. [43] found telomerase activity in male and female organs of tobacco flowers, and Fitzgerald et al. [33] found high levels of activity in young floral buds of cauliflower. It is likely that telomerase activation occurs briefly in the flowering process because telomeres continually shorten during development of the barley inflorescence spike and again during embryogenesis [32]. However, this conclusion is speculative since the temporal and spatial boundaries of telomerase activation during flowering have not been systematically characterized in any species.
Just as we have no clear idea of when telomerase is activated during flowering, we have no idea of when it is repressed during development. Does telomerase remain active throughout embryogenesis? Is it active during germination? Can telomerase really be absent in the shoot apex (particularly if the apex will continue to function for several centuries), or is it confined to just a few meristematic cells?
It is also unknown whether telomerase becomes activated during vegetative reproduction as it does during sexual reproduction. Telomerase expression during vegetative reproduction promises to be an interesting area of research. In vegetative tissues of most plants that have been examined, telomerase activity is absent or barely detectable even with the very sensitive TRAP assay. What happens to telomeres in plants derived from such tissues? Do the telomeres continually shorten, resulting in a Hayflick limit on the rounds of propagation, or is there some mechanism for inducing telomerase or an alternative mechanism for maintaining telomeres? Many horticultural crops have been vegetatively propagated for years without appearing to lose their chromosomes, indicating that telomerase or an alternative mechanism for telomere maintenance is activated during vegetative propagation.
Finally, why should telomerase be deactivated in vegetative tissues? If telomerase is constitutively expressed would telomeres continue to elongate, or is there some mechanism for keeping them within a fixed size range? Are there any consequences to the cell or plant if telomeres are excessively long? The broad range of telomere lengths in maize and the very long telomeres in barley suspension cultures argue that telomeres can carry out their basic functions at almost any length, but the lengthening and shortening of barley telomeres during flowering and embryogenesis suggests the existence of a developmental program for rapidly adjusting the number of telomeric repeats. The purpose of such a system is not clear.
Many of the questions about telomeres and telomerase in plants can be investigated with current technology, especially telomere length measurements and TRAP assays, but for other questions, purification of a plant telomerase and cloning the genes that encode it will be required.
We thank Craig Nessler for critically reading the manuscript and Amanda Neill for drawing Fig. 2. Research on plant telomerases in our laboratories is supported by a grant from the Interdisciplinary Research Initiative of Texas A&M University and by NIH grant GM49157 to D. E. Shippen.
LITERATURE CITED
1.Blackburn, E. H. (1992) Ann. Rev. Biochem.,
61, 113-129.
2.Bacchetti, S. (1996) Cell Dev. Biol.,
7, 31-39.
3.Harley, C. B. (1995) in Telomeres
(Blackburn, E. H., and Greider, C. W., eds.) Cold Spring Harbor
Laboratory Press, Plainview, New York, pp. 247-263.
4.Curry, D. R. (1964) Ecology, 46,
564-566.
5.Lara, A., and Villalba, R. (1993) Science,
260, 1104-1106.
6.Hall, R. D., Riksen-Bruinsma, T., Weyens, G.,
Lefebvre, M., Dunwell, J. M., and Krens, F. A. (1996) Plant
Physiol., 112, 889-892.
7.Wilmut, I., Schnieke, A. E., McWhir, J., Kind, A.
J., and Campbell, K. H. S. (1997) Nature, 385,
810-813.
8.McClintock, B. (1939) Proc. Natl. Acad. Sci.
USA, 25, 405-416.
9.McClintock, B. (1941) Genetics, 26,
234-282.
10.Werner, J. E., Kota, R. S., Gill, B. S., and
Endo, T. R. (1992) Genome, 35, 844-848.
11.Schweizer, L., Yerk-Davis, G. L., Phillips, R.
L., Srienc, F., and Jones, R. J. (1995) Proc. Natl. Acad. Sci.
USA, 92, 7070-7074.
12.Richards, E. J., and Ausubel, F. M. (1988)
Cell, 53, 127-136.
13.Moyzis, R. K., Buckingham, J. M., Cram, L. S.,
Dani, M., Deaven, L. L., Jones, M. D., Meyne, J., Ratliff, R. L., and
Wu, J.-R. (1988) Proc. Natl. Acad Sci. USA, 85,
6622-6626.
14.Pardue, M.-L. (1995) in Telomeres
(Blackburn, E. H., and Greider, C. W., eds.) Cold Spring Harbor
Laboratory Press, Plainview, New York, pp. 339-370.
15.Richards, E. J., Chao, S., Vongs, A., and Yang,
J. (1992) Nucleic Acids Res., 20, 4039-4046.
16.Burr, B., Burr, F. A., Matz, E. C., and
Romero-Severson, J. (1992) Plant Cell, 4, 953-960.
17.Ganal, M. W., Lapitan, N. L. V., and Tanksley, S.
D. (1991) Plant Cell, 3, 87-94.
18.Higashiyama, T., Maki, S., and Yamada, T. (1995)
Mol. Gen. Genet., 246, 29-36.
19.Fuchs, J., Brandes, A., and Schubert, I. (1995)
Pl. Syst. Evol., 196, 227-241.
20.Petracek, M. E., Lefebvre, P. A., Silflow, C. D.,
and Berman, J. (1990) Proc. Natl. Acad. Sci. USA, 87,
8222-8226.
21.Pich, U., Fuchs, J., and Schubert, I. (1996)
Chromosome Res., 4, 207-213.
22.Copenhaver, G. P., and Pikaard, C. S. (1996)
Plant J., 9, 259-272.
23.Bedbrook, J. R., Jones, J., Thompson, R. D., and
Flavell, R. B. (1980) Cell, 19, 545-560.
24.Kilian, A., and Kleinhofs, A. (1992) Mol. Gen.
Genet., 235, 153-156.
25.Fajkus, J., Kovarik, A., Královics, R.,
and Bezdék, M. (1995) Mol. Gen. Genet., 247,
633-638.
26.Zakian, V. A. (1995) Science, 270,
1601-1606.
27.Zentgraf, U. (1995) Plant Mol. Biol.,
27, 467-475.
28.Zhong, Z., Shuie, L., Kaplan, S., and de Lange,
T. (1992) Mol. Cell. Biol., 13, 4834-4843.
29.Shore, D. (1994) Trends Genet.,
10, 408-412.
30.Gray, J. T., Celander, D. W., Price, C. M., and
Cech, T. R. (1991) Cell, 67, 807-824.
31.Bilaud, T., Koering, C. E., Binet-Brasselet, E.,
Ancelin, K., Pollice, A., Gasser, S. M., and Gilson, G. (1996)
Nucleic Acids Res., 24, 1294-1303.
32.Kilian, A., Stiff, C., and Kleinhofs, A.
(1995) Proc. Natl. Acad. Sci. USA, 92, 9555-9559.
33.Fitzgerald, M. S., McKnight, T. D., and Shippen,
D. E. (1996) Proc. Natl. Acad. Sci. USA, 93,
14422-14427.
34.Bucholc, M., and Buchowicz, J. (1995) Plant
Mol. Biol., 27, 435-439.
35.Fajkus, J., Kovarik, A., and Královics, R.
(1996) FEBS Lett., 391, 307-309.
36.Broun, P., Ganal, M. W., and Tanksley, S. D.
(1992) Proc. Natl. Acad. Sci. USA, 89, 1354-1357.
37.Lingner, J., Hughes, T. R., Shevchenko, A., Mann,
M., Lundblad, V., and Cech, T. R. (1997) Science, 276,
561-567.
38.Collins, K., Kobayashi, R., and Greider, C. W.
(1995) Cell, 81, 677-686.
39.Shippen-Lentz, D., and Blackburn, E. H. (1990)
Science, 247, 546-552.
40.Lingner, J., Hendrick, L. L., and Cech, T. R.
(1994) Genes Dev., 8, 1984-1998.
41.Singer, M. S., and Gottschling, D. E. (1994)
Science, 266, 404-409.
42.Kim, N. W., Piatyszek, M. A., Prowse, K. R.,
Harley, C. B., West, M. D., Ho, P. L., Coviello, G. M., Wright, W. E.,
Weinrich, S. L., and Shay, J. W. (1994) Science, 266,
2011-2014.
43.Heller, K., Kilian, A., Piatyszek, M. A., and
Kleinhofs, A. (1996) Mol. Gen. Genet., 52, 342-345.
44.Wang, S., Lapitan, N. L. V., Roder, M., and
Tsuchiya, T. (1992) Genome, 35, 975-980.