Bacteria and Phenoptosis
O. A. Koksharova1,2
1Belozersky Institute of Physico-Chemical Biology, Lomonosov Moscow State University, 119992 Moscow, Russia; fax: +7 (495) 939-3182; E-mail: koksharova@genebee.msu.ru; oa-koksharova@rambler.ru2Institute of Molecular Genetics, Russian Academy of Sciences, pl. Akademika Kurchatova 2, 123182 Moscow, Russia; fax: +7 (499) 196-0221
Received May 28, 2013
Genetically programmed death of an organism, or phenoptosis, can be found not only in animals and plants, but also in bacteria. Taking into account intrapopulational relations identified in bacteria, it is easy to imagine the importance of phenoptosis in the regulation of a multicellular bacterial community in the real world of its existence. For example, autolysis of part of the population limits the spread of viral infection. Destruction of cells with damaged DNA contributes to the maintenance of low level of mutations. Phenoptosis can facilitate the exchange of genetic information in a bacterial population as a result of release of DNA from lysed cells. Bacteria use a special “language” to transmit signals in a population; it is used for coordinated regulation of gene expression. This special type of regulation of bacterial gene expression is usually active at high densities of bacteria populations, and it was named “quorum sensing” (QS). Different molecules can be used for signaling purposes. Phenoptosis, which is carried out by toxin–antitoxin systems, was found to depend on the density of the population; it requires a QS factor, which is called the extracellular death factor. The study of phenoptosis in bacteria is of great practical importance. The components that make up the systems ensuring the programmed cell death, including QS factor, may be used for the development of drugs that will activate mechanisms of phenoptosis and promote the destruction of pathogenic bacteria. Comparative genomic analysis revealed that the genes encoding several key enzymes involved in apoptosis of eukaryotes, such as paracaspases and metacaspases, apoptotic ATPases, proteins containing NACHT leucine-rich repeat, and proteases similar to mitochondrial HtrA-like protease, have homologs in bacteria. Proteomics techniques have allowed for the first time to identify the proteins formed during phenoptosis that participate in orderly liquidation of Streptomyces coelicolor and Escherichia coli cells. Among these proteins enzymes have been found that are involved in the degradation of cellular macromolecules, regulatory proteins, and stress-induced proteins. Future studies involving methods of biochemistry, genetics, genomics, proteomics, transcriptomics, and metabolomics should support a better understanding of the “mystery” of bacterial programmed cell death; this knowledge might be used to control bacterial populations.
KEY WORDS: bacteria, phenoptosis, programmed cell death, cell population, QS, autoinducers, extracellular death factor, comparative genomics, proteomicsDOI: 10.1134/S0006297913090010
Abbreviations: ALD, apoptotic-like death; EDF, extracellular death factor; PCD, programmed cell death; QS, quorum sensing; TA complex, toxin–antitoxin complex.
Mankind has been interested in issues concerning lifespan of organisms
on our planet since ancient times. The early physician Claudius Galen
(II century AD) introduced the term “apoptosis”, which
means “leaf fall” in Greek. Galen used this term to
describe the process leading to the falling of leaves in autumn. As the
leaves fall only from live trees, the scientist concluded that this
property is incorporated in the program of development of these plants.
The hypothesis of the programmed death of organisms was first proposed
by German scholar August Weismann in the 1880s. According to this
hypothesis, a genetically programmed mechanism of cell death appeared
as a result of natural selection so as to eliminate old worn-out
individuals, thus freeing living space and resources for younger
generations. In the late 1990s, V. P. Skulachev suggested the term
“phenoptosis” to define the programmed death of an organism
[1-4]. All the properties of an
organism are encoded in its genome, including the processes of dying,
and they are realized in the form of a chain of biochemical reactions
that ultimately cause its death. That is why the phenomenon of
phenoptosis is actively studied by biochemists. Geneticists, molecular
biologists, and bioinformaticists have joined the study of this process
in the last ten to fifteen years. Data arrays on the genomes of
different organisms have become available. Analysis of these data as
well as their use for proteomic, transcriptomic, and metabolomic
studies can facilitate better understanding of the mechanisms of aging
and programmed death of organisms, their regulation, and evolution.
Mechanisms and evolution of apoptosis in unicellular organisms have been discussed earlier in the Russian literature reviews [5-7]. This review is dedicated to the coverage of some new facts obtained in recent years by researchers working in this field, and to the review of experimental data indicating the presence of “communication” between prokaryotic cells that can lead to bacterial phenoptosis.
BACTERIA IN A POPULATION ARE AN ANALOG OF A MULTICELLULAR
ORGANISM
Bacteria are known not to live individually in nature or in the laboratory. Bacteria form colonies (progeny of a single cell) and biofilms, i.e. they exist in the form of cell populations. Contemporary research supports the idea that bacterial populations can be seen as holistic formations with the division of biochemical functions between the members of the microbial community, which makes these populations somewhat analogous to multicellular organisms. The motto “One for all and all for one!” can be applied also to cells in bacterial populations. Bacterial “altruism” finds experimental support. For example, there exists a known problem of the resistance of bacterial populations to antibiotics. Recent studies show that bacterial “altruism” may play an important role in this resistance. Lee et al. [8] showed the majority of individual bacteria of Escherichia coli in a population exhibiting resistance to antibiotic norfloxacin to be significantly more sensitive to this antibiotic compared to the general population. This phenomenon could be explained by the fact that only a small number of highly resistant mutant bacteria in the population “altruistically” protected more vulnerable cells, in particular, forming indole – a signaling molecule normally produced by actively growing cells that do not experience stress. In this case, it supported the growth of the entire bacterial population in stressful conditions created by the presence of the antibiotic in the medium. The diversity of bacterial phenotypes in one population allows them to adapt to the constantly changing environmental conditions, which, in turn, determine the need for certain phenotypes [9, 10].
Bacteria use a special “language” to transmit signals in a population; it is used for coordinated regulation of gene expression. This type of regulation of bacterial gene expression, which usually functions at high densities of bacterial populations, is called “quorum sensing” (QS). QS systems are characterized by having at least two main components: low molecular mass signaling molecules (autoinducers) of varying nature, which diffuse from the cells to the medium and back, and receptor regulatory proteins that interact with signaling molecules. Signaling molecules of the QS systems facilitate intercellular communication in bacterial populations, providing coordinated response to changes in environmental conditions.
Different molecules can be used for signaling purposes. Several classes of signaling molecules are now known: oligopeptides in Gram-positive bacteria and N-acyl homoserine lactones (AHL, AI-1) in Gram-negative bacteria. Furthermore, there is a family of autoinducers known as autoinducers-2 (AI-2) both in Gram-negative and Gram-positive bacteria [11-15]. They are derivatives of 2-methyl-2,3,4,5-tetra-oxytetrahydrofuran (AI-2, THMF). There is also another group of regulatory molecules – compounds related to adrenaline (AI-3) and derivatives of indole and quinoline [15]. Signaling molecules are used both for communication between bacteria of one species and for interspecies communication. Therefore, suppression of communication between bacterial cells by blocking signaling molecules is seen as a new approach in the treatment of infections [12, 15].
Apoptosis or programmed cell death (PCD) is a genetic program of cell death in multicellular eukaryotic organisms. It contributes to the preservation of order and normal functioning of a biological system by cleaning it from unneeded and damaged cells, cells that have completed their life cycle, and those potentially dangerous cells that have resulted from mutations.
PCD was long believed to be characteristic only for multicellular animals and plants. At first glance, apoptosis seems to be useless for unicellular organisms. Under ideal growth conditions (continuous supply of nutrients, removal of metabolic products, lack of stress) bacteria could live forever: each cell would produce two new cells resulting from division, and this could go on forever. However, in an actual habitat apoptosis may play an important role in the regulation of a bacterial community. For example, autolysis of a part of a population can limit the spread of viral infection. The destruction of cells with damaged DNA will help to maintain a low level of mutations. PCD can facilitate the exchange of genetic information in bacterial population as a result of DNA release from lysed cells. One can call this phenomenon “altruism” of dying bacterial cells [16] – “die yourself, but save your comrades”. Some of the cells in the population trigger a mechanism leading to cell death, which results in release of compounds used by the remaining cells [6]. The ability for “horizontal gene transfer”, which occurs as a result of lysis of individual cells, is an important factor in bacterial evolution. The released bacterial plasmid and genomic DNA can be taken up by other cells of the population, this process leading to the transfer of genes changed by mutations. This may result in the acquisition of new properties by bacterial cells and their further evolution.
MECHANISMS OF BACTERIAL APOPTOSIS AND QS
“Toxin–antitoxin” systems. PCD genetic mechanisms are not fully understood. Much attention has been focused on study of “toxin–antitoxin” systems (TA systems) found in E. coli and many other bacteria, including pathogens [17-19]. TA systems are divided into three classes – I, II, and III – based on the nature of the antitoxin and its mode of action. Small noncoding RNA molecules are antitoxins operating in systems I and III. In type I systems, expression of the toxin-encoding gene is inhibited by antisense RNA transcribed in the opposite direction [20, 21]. In type III systems, the antitoxin is an RNA molecule that interacts with toxin protein and inhibits its activity [22]. Antitoxins of type II are proteins that form toxin–antitoxin complexes (TA complexes) with no toxic activity [23].
Identified TA toxins belong to different protein families with different biochemical activities [23-25]. The vast majority of toxins of this type are specific endoribonucleases (mRNA interferases), e.g. the MazF, RelE, and HicA toxins [24]. They form TA modules composed of a pair of genes in a bacterial chromosome: a gene encoding a stable toxin, which causes cell death, and a gene encoding a labile antitoxin, which counteracts the activity of the toxin.
MazEF system of E. coli. The mazEF system of E. coli is one of the best studied. It was first described as a mechanism responsible for bacterial PCD [26-28]. The mazF gene encodes a stable cytotoxic protein, and mazE encodes an unstable antidote to MazF protein, which is quickly degraded by the ATP-dependent ClpAP serine protease [26]. Under balanced growth conditions, as long as MazE and MazF are coexpressed, MazE neutralizes the toxic effect of MazF [26], i.e. the toxin is constantly neutralized by the antitoxin. The MazF toxin is a “long-lived” protein, an endoribonuclease, which cleaves RNA in ACA sequences [29, 30]. However, under stress conditions (starvation, effects of antibiotics, reactive oxygen species, presence of 3′,5′-bispyrophosphate (ppGpp), DNA damage), which prevent expression of the mazEF operon, the equilibrium is disturbed, and the toxin “poisons” the cell, destroying the cellular mRNA, which leads to cell death and autolysis of the greater part of the population [26, 31]. Under stressful conditions, the labile protein antitoxin MazE is destroyed by the ClpAP protease, and as a result the more stable MazF toxin is free to act, ultimately causing cell death.
PCD systems were first discovered mainly in E. coli cells in low-copy plasmids. When a bacterium loses such a plasmid in the course of division, it no longer contains the “toxin–antitoxin” operon, and after the rapid antitoxin inactivation it will be killed by the toxin remaining in the cell [32, 33]. Genes encoding the components of a PCD system have now been found in genomic chromosomal DNA of many free-living bacteria [34-36]. This system is believed to help bacteria adapt to stressful environmental conditions. According to this hypothesis, under adverse conditions the antitoxin will be destroyed by proteases, their activity being induced by stress. In the absence of antitoxin, the TA operon will be transcribed and the toxin will be released from the TA complex. Consequently, the released toxins will cause growth cessation and cell death by inhibiting such important cellular processes as protein synthesis and DNA replication [32, 37]. It should be noted that successive removal of all ten TA systems encoding endoribonuclease toxins in E. coli has a cumulative effect and leads to the formation of a subpopulation of bacterial cells with a low level of metabolism and low resistance to antibiotics [38], suggesting functional redundancy of TA systems in bacterial cells.
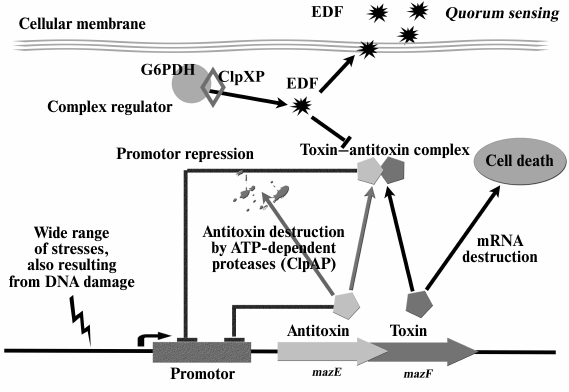
MazEF-mediated apoptosis was found to be a phenomenon depending on population density that requires a QS factor called extracellular death factor (EDF) [39-41]. Structural analysis has shown EDF to be a linear pentapeptide, Asn-Asn-Trp-Asn-Asn. Each of the five EDF amino acids is essential to its activity [39]. The mazEF module as well as glucose-6-phosphate dehydrogenase (G6PD, zwf gene) and ClpXP protease were shown to be essential for synthesis of EDF [40]. This peptide is a small fragment of the G6PD enzyme. The pentapeptide is cut out from the segment between catalytic and structural domains of this enzyme. The search for similar nucleotide sequences in the genome revealed only five open reading frames, parts of which could encode EDF. However, only the deletions of the zwf gene and the yeo gene of unknown function were shown to prevent the formation of an active extracellular death factor [39].
There is a positive feedback between the mazEF module and EDF activity – the latter is required for MazF activation under all studied stress conditions. At the same time, MazF activation resulted in increased EDF synthesis, which, in turn, caused increased cell death [40]. Thus, EDF is an integral part of the mazEF system (Fig. 1).
Fig. 1. The mazEF system of E. coli and extracellular death factor (EDF). Under stress, labile antitoxin MazE is destroyed by ClpAP protease, and the more stable MazF toxin causes cell death. Activity of stress-induced ClpXP protease leads to the formation of EDF peptide, which is a QS factor inhibiting the formation of MazEF complex.
ChpBI–ChpBK TA system in E. coli. The QS factor EDF has recently been discovered to participate in the functioning of yet another TA system in E. coli, ChpBI–ChpBK [42]. ChpBK is a toxin homologous to MazF; it is a site-specific endoribonuclease that splits a single-stranded RNA in the sequences ACA, ACG, and ACU [43]. The signaling EDF molecule was found to significantly enhance endoribonuclease activity of both the MazF and the ChpBK proteins in an in vitro system. EDF also neutralizes the inhibitory effect of MazE antitoxin on MazF toxin and of ChpBI antitoxin on ChpBK toxin. Furthermore, EDF was shown to bind directly and site-specifically with MazF [42]. These data are of great interest as so far signaling molecules belonging to the systems of QS regulation have been known to control gene expression at the level of transcription, while EDF was believed to act post-transcriptionally. It should also be noted that EDF is the only peptide-type signaling molecule found in E. coli.
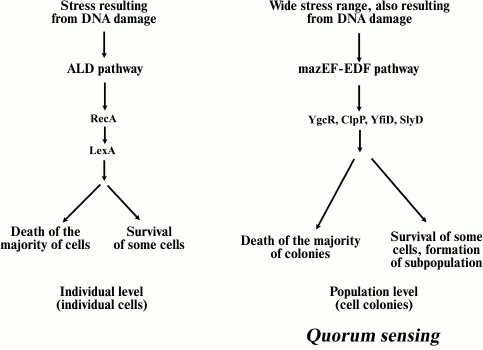
Apoptotic-like death. Extensive DNA damage can lead to cell death following a second pathway called apoptotic-like death (ALD) [44]. ALD is similar to apoptosis of eukaryotic cells. The cell membrane is depolarized and DNA is fragmented in the course of this process. Erental et al. have shown ALD to result from the activity of two proteins, RecA and LexA [44]. LexA protein is an inhibitor of the SOS response, which is a global response of a bacterial cell to DNA damage, when the cell cycle is interrupted and DNA reparation is induced. Thus, ALD can be defined as a form of SOS response. Those authors concluded that in the case of E. coli cells, the ALD pathway is a backup system in relation to mazEF, the main pathway of cell death [44]. If some components of the mazEF pathway are inactivated, bacterial cell death will follow the ALD pathway (Fig. 2). In this case RecA inactivates LexA, which is a repressor of the transcription of SOS-system genes. Essentially, the destruction of LexA protein activates the SOS response. The mazEF system-mediated mechanism of cell death also inhibits the ALD pathway by reducing the mRNA level of RecA protein.
Fig. 2. Two pathways of bacterial PCD. The ALD pathway is induced only by stress caused by DNA damage, and it acts only at the level of individual cells. The mazEF pathway mechanism is initiated by different stress types and acts at the level of a bacterial population, involving also QS regulation. This pathway leads to the death of most bacteria and the formation of a small surviving cell subpopulation, which will give rise to a new bacterial population. The YgcR, ClpP, YfiD, and SlyD proteins are involved in bacterial cell phenoptosis.
So, what is the evolutionary significance of these two pathways for bacteria? The mazEF pathway provides for the survival of a small subpopulation of cells under stress. Surviving cells will give rise to a new population as soon as conditions become optimal. Therefore, the mazEF pathway of cell death can be called an “altruistic” mechanism of bacterial survival under stress. Conversely, the ALD pathway may operate at the level of individual cells (in contrast to a cell population), since survival results from reparation of individual damaged cells. This pathway may serve as a backup as the ALD mechanism responds only to stress caused by DNA damage, but not to other stresses, and surviving cells may be less sensitive to other types of stress conditions. We still need to discover the evolutionary connection between these pathways.
The concept of PCD in bacteria forces us to reconsider a wide range of important phenomena in the life of a microbial cell, such as cell properties in a steady state, the nature of resistance to antibiotics and other stress factors, and related issues of the survival of bacteria in biofilms. Study of the mechanisms of bacterial apoptosis is of great practical importance. Components of PCD, including QS factor, may be used for the development of drugs that will activate PCD, thus promoting the destruction of pathogens in the course of antibiotic treatment [45].
ORIGIN OF PHENOPTOSIS: GENOMIC AND PROTEOMIC STUDIES
Comparative genomic analysis. Availability of complete genomic nucleotide sequences of prokaryotes and data on eukaryotic genomes has made possible comparative genomic analysis that sheds light on the origin of apoptosis in bacteria. For example, some key enzymes involved in apoptosis in eukaryotes (such as enzymes belonging to the families of paracaspases and metacaspases, which, in turn, are part of a superfamily of caspase-like proteases, apoptotic ATPases, NACHT-repeat domain-containing proteins, and proteases similar to mitochondrial HtrA protease) have homologs in bacteria [46]. Computer analysis of the structure and configuration of the homologs of apoptotic proteins, in particular, apoptotic ATPases and caspases, suggests the possibility of formation of large protein complexes in bacteria of sophisticated development and differentiation, such as actinomycetes, cyanobacteria, myxobacteria, and some α-proteobacteria. These protein complexes can be functional analogs or perhaps evolutionary precursors of eukaryotic apoptosomes [46]. It should be noted that so far there have not been enough experimental studies of such complexes, including their possible role in signal transmission in bacteria.
An interesting hypothesis on the evolutionary development of PCD mechanisms was introduced in the work of Frade and Michaelidis [47]. Ancient endosymbionts represented by α-proteobacteria could use secreted and membrane proteases such as metacaspases, paracaspases, and HtrA-like proteases to kill their host cell if for some reason it became “inhospitable” for the endosymbiont, for example, due to the lack of nutrients. Such a mechanism would allow the endosymbiont to leave the host cell and move into another host. In the course of subsequent evolution, this “aggression” mechanism could have subdued to the host and gradually could have changed into the PCD mechanism including regulatory components [46].
Understanding of PCD mechanisms in prokaryotes is important for solving many practical problems. These include bacterial infection of humans and animals [15] and problems of purity and safety of aquatic ecosystems [48]. In the last decade, increasing attention has been given to problems of aquatic ecosystems, “bloom” of phytoplankton because of secreted toxins. So far there have not been many experimental works that could help understand the regulation of relationships in such photoautotrophic populations of microorganisms. Expanding, phototrophic bacterial blooms can unexpectedly quickly “disappear”, become lysed. What triggers the simultaneous death of all the cells in the population? Phytoplankton has a core group of proteins that are orthologs of caspases of multicellular animals. It is assumed that PCD in prokaryotic plankton and in independently evolved eukaryotic lines may have common evolutionary roots [49].
Typically, cell death in prokaryotic and eukaryotic unicellular phytoplankton is caused by environmental stress factors (for example, starvation, high-intensity light, oxidative stress, UV irradiation) [49-53]. However, the rate of cell lysis in such a population is very high, and it cannot be explained by viral attack or zooplankton rapidly eating phytoplankton. This cellular self-destruction is similar to PCD of multicellular organisms, a form of suicide triggered by the cell, when an endogenous biochemical pathway leads to morphological changes characteristic of apoptosis and, ultimately, to cell dissolution.
The PCD mechanism includes biochemical coordination of specialized multicomponent cell machinery consisting of receptors, adaptors, signaling kinases, and proteases. A specific class of intracellular cysteine proteases that split proteins solely after aspartate is called caspases (caspase; cysteine-dependent aspartate specific protease). These proteases are of particular interest since they participate both in initiation and implementation of PCD by cutting different proteins essential for the cell in response to proapoptotic signals [54]. Discovery of homologs of caspases, paracaspases, and metacaspases in various organisms, including animals, higher plants, unicellular protozoa, fungi, and bacteria [46, 55], suggest that they may represent ancestral enzymes that led to the development of cell death mechanisms. The earliest ancestors of plants and animals are likely to have had a minimal set of components involved in apoptosis. More complex systems have evolved along with the emergence of multicellular organisms. Since the majority of metacaspases have been characterized only in silico, they were correlated to the known caspases of yeast [56] and trypanosomes [57]. However, metacaspases (along with other proteins involved in PCD) proved to be widespread in the genomes of prokaryotic and eukaryotic planktonic organisms [49]. Moreover, morphological and biochemical characteristics of caspase-mediated PCD could be observed in cells of different representatives of phytoplankton, including cyanobacteria, green algae, and dinoflagellates [50-53].
The PCD phenomenon has been studied using different bacterial models: Helicobacter [58], Anabaena [59], Xanthomonas [60], Staphylococcus aureus [61], Streptomyces [62]. It was found that bacteria with complex development mechanisms, such as Streptomyces, Bacillus, Anabaena, Caulobacter, Rhizobium, and Myxobacteria, can trigger PCD; these bacteria have genes encoding proteins that are phylogenetically related to the proteins involved in apoptosis in eukaryotes [46, 63].
Proteomics of phenoptosis. Proteomic studies of bacterial PCD are still very few. Methods of proteomics have allowed for the first time to identify proteins formed during PCD processes that are involved in the orderly liquidation of Streptomyces coelicolor cells. Enzymes involved in degradation of cellular macromolecules, regulatory and stress-induced proteins were found among these proteins [64]. Enzymes participating in metabolism of fatty acids, which are presumably involved in membrane degradation, different hydrolases, and proteases were shown to be active during PCD. In particular, ATP-binding subunit of C complex constituting the Clp protease has been identified. This protease is known to possess the properties of a global regulator; it is associated with QS regulation [65, 66] and is often manifested under stress conditions in bacteria. In this study, AAA-ATPase, kinases, chaperones, and also proteins whose biological role in PCD is still difficult to explain were identified [64]. Among this group a number of proteins was discovered: glyoxylate carboligase, glycosyltransferase, various forms of DNA methylase, two acetyl transferases involved in biosynthesis of siderophores, and hypothetical proteins whose existence had been predicted earlier only at the level of genome annotation, while proteomic research showed them to be actually synthesized in cells, and furthermore they were shown to be related to PCD.
Another proteomic study [67] showed that although MazF toxin induction in E. coli cells leads to the inhibition of the synthesis of most cellular proteins, the synthesis of a special group of small proteins (less than 20 kDa) still takes place. Some of these have been identified, and these proteins were shown to be required for the destruction of the main population of bacterial cells. ClpP protease, SlyD, YfiD, ElaC, YgcR, and YfbU proteins were among the identified proteins. In this study proteins with the opposite function were also found – “survival” proteins needed for the maintenance of a small part of the cellular subpopulation (YajQ, RsuA, DeoC, SoxR, and SoxS). Thus, MazF is likely to function as a regulator triggering the processes leading to the death of most of the population and to the survival of its small part, which would become a “core” of the new bacterial population when conditions become more favorable.
Such proteomic studies are very important for a deeper understanding of PCD mechanisms and regulation; they open up new perspectives for genetic, phylogenetic, and functional studies of bacterial phenoptosis.
The accumulated experimental data suggest prokaryotic cells to be not that “simple” in regulation of their metabolism, cell division, “social” behavior, and death. The presence of eukaryotic analogs of tubulin and actin in bacteria; recognition of the fact that prokaryotic cells possess cytoskeleton; discovery of circadian rhythms in cyanobacterial cells; the ability for cell differentiation present in some bacteria; discovery of a variety of signaling molecules controlling the behavior of microbial populations contributed to the formation of a new view of microorganisms. Bacteria are shown to possess many characteristics inherent in eukaryotic cells; in particular, they were proved capable of phenoptosis. Future studies involving methods of mutagenesis, genomics, proteomics, transcriptomics, and metabolomics will facilitate better understanding of the “mystery” of bacterial programmed death and the use of this knowledge to control bacterial populations.
This work was supported in part by the Russian Foundation for Basic Research (grant 11-04-00774).
REFERENCES
1.Skulachev, V. P. (1997) Biochemistry
(Moscow), 62, 1191-1195.
2.Skulachev, V. P. (1999) Biochemistry
(Moscow), 64, 1418-1426.
3.Longo, V. D., Mitteldorf, J., and Skulachev, V. P.
(2005) Nat. Rev. Genet., 6, 886-872.
4.Skulachev, V. P. (2012) Biochemistry
(Moscow), 77, 689-706.
5.Oleskin, A. V. (2001) Soros Obrazovat. Zh.,
8, 7-12.
6.Gordeeva, A. V., Labas, Y. A., and Zvyagilskaya, R.
A. (2004) Biochemistry (Moscow), 69, 1055-1066.
7.Prozorov, A. A., and Danilenko, V. N. (2010)
Microbiology (Moscow), 79, 129-140.
8.Lee, H. H., Molla, M. N., Cantor, C. R., and
Collins, J. J. (2010) Nature, 467, 82-85.
9.Tanouchi, Y., Pai, A., Buchler, N. E., and You, L.
(2012) Mol. Syst. Biol., 8, 626.
10.Reuven, P., and Avigdor, E. (2011) Curr. Opin.
Genet. Dev., 21, 759-767.
11.Khmel, I. A. (2006) Microbiology (Moscow),
75, 390-397.
12.Khmel, I. A., and Metlitskaya, A. Z. (2006)
Mol. Biol. (Moscow), 40, 169-182.
13.Waters, C., and Bassler, B. (2005) Annu. Rev.
Cell Dev. Biol., 21, 319-346.
14.Miller, M. B., and Bassler, B. L. (2001) Annu.
Rev. Microbiol., 55, 165-199.
15.Shpakov, A. O. (2009) Microbiology
(Moscow), 78, 133-143.
16.Carmona-Fontaine, C., and Xavier, J. B. (2012)
Mol. Syst. Biol., 8, 627-628.
17.Mittenhuber, G. (1999) J. Mol. Microbiol.
Biotechnol., 1, 295-302.
18.Hayes, F. (2003) Science, 301,
1496-1499.
19.Pandey, D. P., and Gerdes, K. (2005) Nucleic
Acids Res., 33, 966-976.
20.Gerdes, K., and Wagner, E. G. (2007) Curr.
Opin. Microbiol., 10, 117-124.
21.Fozo, E. M., Hemm, M. R., and Storz, G. (2008)
Microbiol. Mol. Biol. Rev., 72, 579-589.
22.Fineran, P. C., Blower, T. R., Foulds, I. J.,
Humphreys, D. P., Lilley, K. S., and Salmond, G. P. (2009) Proc.
Natl. Acad. Sci. USA, 106, 894-899.
23.Gerdes, K., Christensen, S. K., and
Lobner-Olesen, A. (2005) Nat. Rev. Microbiol., 3,
371-382.
24.Yamaguchi, Y., Park, J. H., and Inouye, M. (2011)
Annu. Rev. Genet., 45, 61-79.
25.Mutschler, H., Gebhardt, M., Shoeman, R. L., and
Meinhart, A. (2011) PLoS Biol., 9, e1001033.
26.Aizenman, E., Engelberg-Kulka, H., and Glaser, G.
(1996) Proc. Natl. Acad. Sci. USA, 93, 6059-6063.
27.Mittenhuber, G. (1999) J. Mol. Microbiol.
Biotechnol., 1, 295-302.
28.Engelberg-Kulka, H., Hazan, R., and Amitai, S.
(2005) J. Cell Sci., 118, 4327-4332.
29.Zhang, Y., Zhang, J., Hoeflich, K. P., Ikura, M.,
Qing, G., and Inouye, M. (2003) Mol. Cell, 12,
913-923.
30.Zhang, Y., Zhang, J., Hara, H., Kato, I., and
Inouye, M. (2005) J. Biol. Chem., 280, 3143-3150.
31.Hazan, R., Sat, B., and Engelberg-Kulka, H.
(2004) J. Bacteriol., 186, 3663-3669.
32.Hayes, F. (2003) Science, 301,
1496-1499.
33.Gerdes, K., Christensen, S. K., and
Lobner-Olesen, A. (2005) Nat. Rev. Microbiol., 3,
371-382.
34.Pandey, D. P., and Gerdes, K. (2005) Nucleic
Acids Res., 33, 966-976.
35.Makarova, K., Wolf, Y., and Koonin, E. (2009)
Biol. Direct., 4, 19.
36.Leplae, R., Geeraerts, D., Hallez, R.,
Guglielmini, J., Dreze, P., and van Melderen, L. (2011) Nucleic
Acids Res., 39, 5513-5525.
37.Yamaguchi, Y., and Inouye, M. (2011) Nat. Rev.
Microbiol., 9, 779-790.
38.Maisonneuve, E., Shakespeare, L. J., Jorgensen,
M. G., and Gerdes, K. (2011) Proc. Natl. Acad. Sci. USA,
108, 13206-13211.
39.Kolodkin-Gal, I., Hazan, R., Gaathon, A.,
Carmeli, S., and Engelberg-Kulka, H. (2007) Science, 318,
652-655.
40.Kolodkin-Gal, I., and Engelberg-Kulka, H. (2008)
J. Bacteriol., 190, 3169-3175.
41.Amitai, S., Kolodkin-Gal, I., Hananya-Meltabashi,
M., Sacher, A., and Engelberg-Kulka, H. (2009) PLoS Genet.,
5, e1000390; doi: 10.1371/journal.pgen.1000390.
42.Belitsky, M., Avshalom, H., Erental, A., Yelin,
I., Kumar, S., London, N., Sperber, M., Schueler-Furman, O., and
Engelberg-Kulka, H. (2011) Mol. Cell, 41, 625-635.
43.Zhang, Y., Zhu, L., Zhang, J., and Inouye, M.
(2005) J. Biol. Chem., 280, 26080-26088.
44.Erental, A., Sharon, I., and Engelberg-Kulka, H.
(2012) PLoS Biol., 10, e1001281; doi:
10.1371/journal.pbio.1001281.
45.Engelberg-Kulka, H., Sat, B., Reches, M., Amitai,
S., and Hazan, R. (2004) Trends Microbiol., 12,
66-71.
46.Koonin, E. V., and Aravind, L. (2002) Cell
Death Differ., 9, 394-404.
47.Frade, J. M., and Michaelidis, T. M. (1997)
Bioessays, 19, 827-832.
48.Koksharova, O. A. (2010) Microbiology,
79, 721-734.
49.Bidle, K. D., and Falkowski, P. G. (2004) Nat.
Rev. Microbiol., 2, 643-655.
50.Berman-Frank, I., Bidle, K., Haramaty, L., and
Falkowski, P. (2004) Limnol. Oceanogr., 49,
997-1005.
51.Moharikar, S., D’Souza, J. S., Kulkarni, A.
B., and Rao, B. J. (2006) J. Phycol., 42,
423-433.
52.Segovia, M., Haramaty, L., Berges, J. A., and
Falkowski, P. G. (2003) Plant Physiol., 132, 99-105.
53.Vardi, A., Berman-Frank, I., Rozenberg, T.,
Hadas, O., Kaplan, A., and Levine, A. (1999) Curr. Biol.,
9, 1061-1064.
54.Thornberry, N. A., and Lazebnik, Y. (1998)
Science, 281, 1312-1316.
55.Uren, A. G., O’Rourke, K., Pisabarro, M.
T., Seshagiri, S., Koonin, E. V., and Dixit, V. M. (2000) Mol.
Cell, 6, 961-967.
56.Madeo, F., Herker, E., Maldener, C., Wissing, S.,
Lachelt, S., Herlan, M., Fehr, M., Lauber, K., Sigrist, S. J.,
Wesselborg, S., and Frohlich, K.-U. (2002) Mol. Cell, 9,
911-917.
57.Szallies, A., Kubata, B. K., and Duszenko, M.
(2002) FEBS Lett., 517, 144-150.
58.Kusters, J. G., Gerrits, M. M., van Strijp, J.
A., and Vandenbroucke-Grauls, C. M. (1997) Infect. Immun.,
65, 3672-3679.
59.Ning, S. B., Guo, H. L., Wang, L., and Song, Y.
C. (2002) J. Appl. Microbiol., 93, 15-28.
60.Gautam, S., and Sharma, A. (2002) Mol.
Microbiol., 44, 393-401.
61.Bayles, K. W. (2003) Trends Microbiol.,
11, 306-311.
62.Manteca, A., Fernandez, M., and Sanchez, J.
(2006) Res. Microbiol., 157, 143-152.
63.Leipe, D. D., Koonin, E. V., and Aravind, L.
(2004) J. Mol. Biol., 343, 1-28.
64.Manteca, A., Mader, U., Connolly, B. A., and
Sanchez, J. (2006) Proteomics, 6, 6008-6022.
65.He, Y. W., and Zhang, L. H. (2008) FEMS
Microbiol. Rev., 32, 842-857.
66.Tao, F., He, Y. W., Wu, D. H., Swarup, S., and
Zhang, L. H. (2010) J. Bacteriol., 192, 1020-1029.
67.Amitai, S., Kolodkin-Gal, I., Hananya-Meltabashi,
M., Sacher, A., and Engelberg-Kulka, H. (2009) PLoS Genet.,
5, e1000390.