REVIEW: Iron-Sulfur Clusters in Mitochondrial Metabolism: Multifaceted Roles of a Simple Cofactor
Johnny Stiban1*, Minyoung So2, and Laurie S. Kaguni2*
1Birzeit University, Department of Biology and Biochemistry, P.O. Box 14, West Bank 627 Birzeit, Palestine; E-mail: jstiban@birzeit.edu2Michigan State University, Department of Biochemistry and Molecular Biology and Center for Mitochondrial Science and Medicine, East Lansing, 48824 Michigan, USA; E-mail: lskaguni@msu.edu
* To whom correspondence should be addressed.
Received April 1, 2016; Revision received May 10, 2016
Iron-sulfur metabolism is essential for cellular function and is a key process in mitochondria. In this review, we focus on the structure and assembly of mitochondrial iron-sulfur clusters and their roles in various metabolic processes that occur in mitochondria. Iron-sulfur clusters are crucial in mitochondrial respiration, in which they are required for the assembly, stability, and function of respiratory complexes I, II, and III. They also serve important functions in the citric acid cycle, DNA metabolism, and apoptosis. Whereas the identification of iron-sulfur containing proteins and their roles in numerous aspects of cellular function has been a long-standing research area, that in mitochondria is comparatively recent, and it is likely that their roles within mitochondria have been only partially revealed. We review the status of the field and provide examples of other cellular iron-sulfur proteins to highlight their multifarious roles.
KEY WORDS: iron-sulfur clusters, iron-sulfur metabolism, mitochondria, respiration, citric acid cycle, DNA metabolismDOI: 10.1134/S0006297916100059
Abbreviations: BN-PAGE, blue native polyacrylamide gel electrophoresis; cI-III, respiratory complexes I-III; CIA, cytosolic iron-sulfur protein assembly; ETC, electron transport chain; Fe-S, iron-sulfur; IRE, iron-responsive elements; IRP, iron-regulatory protein; ISCs, iron-sulfur clusters; ISP, iron-sulfur protein; mtDNA, mitochondrial DNA; Q, ubiquinone.
Iron is an omnipresent element on earth, and it is the second most
abundant metal [1]. Iron participates in a wide
range of biological cofactors and prosthetic groups. Iron can be
coordinated to amino acid side chains directly (e.g. transferrins), or
via porphyrin rings (e.g. hemoglobins) or sulfur atoms (such as in
iron-sulfur proteins). In this review, we focus on iron-sulfur (Fe-S)
proteins in mitochondrial metabolism.
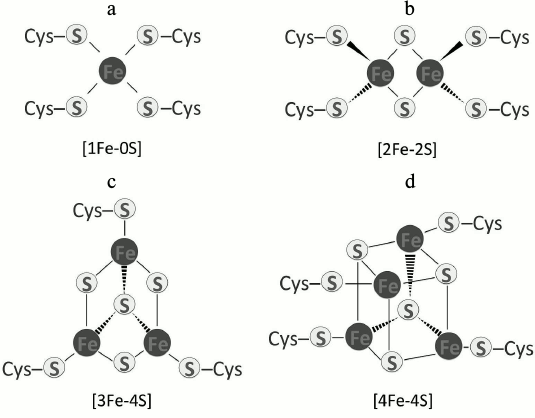
The presence of Fe-S clusters (ISCs) in proteins emerged as life on earth began. It is suggested that ISCs were the first catalysts and enzyme cofactors for many biochemical reactions in the anaerobic world [2]. Their versatility and robustness can be attributed to the extraordinary properties of iron and sulfur atoms because both can readily donate or accept electrons [3-6]. There are many types of ISCs ranging from a single iron atom coordinated to four cysteine sulfhydryl groups [1Fe-0S], such as in rubredoxin [7-9], to more complex types. ISCs such as [2Fe-2S], [3Fe-4S], and [4Fe-4S] predominate (Fig. 1). Other complex clusters also exist, such as [7Fe-8S], [8Fe-7S], or [8Fe-8S] in molybdenum-iron (MoFe) proteins [10]. In most Fe-S proteins, the cluster is coordinated to four sulfhydryl groups in cysteines. Rieske Fe-S proteins use two histidines and two cysteines instead of four cysteines for coordination [11, 12]. Other permutations of coordinating amino acids occur in some Fe-S proteins [13], and in these cases, the chemical nature of the structure is altered. Nonetheless, the basic chemical reactions in which clusters are involved remain the same.
ISCs undergo a variety of reactions and can be converted from one type to another within the course of a reaction. In Fe-S proteins, the iron atom is the donor and acceptor of electrons and it alternates between the oxidized (Fe3+) and reduced (Fe2+) states by the addition or loss of a single electron, generally without the involvement of protons [14]. In vivo, ISCs are incorporated into proteins by a complex pathway requiring numerous accessory proteins [15-17]. The loss or malfunction of these can lead to a wide-range of diseases (at least ten known human genetic diseases) and may ultimately cause death [18-21]. Here, we discuss the various facets of ISC biochemistry in mitochondrial metabolism.
Fig. 1. Fe-S centers that are common in proteins: a) [1Fe-0S]; b) [2Fe-2S]; c) [3Fe-4S]; d) [4Fe-4S].
STRUCTURE AND ASSEMBLY OF MITOCHONDRIAL Fe-S CLUSTERS
Mitochondria share many ancestral pathways with bacteria. Among them is the Iron Sulfur Cluster (ISC) pathway of Fe-S biosynthesis [22]. Eukaryotes developed a complementary system termed the Cytosolic Iron-sulfur protein Assembly (CIA) pathway that inserts Fe-S clusters into a variety of proteins destined for the cytoplasm or nucleus [16, 23].
Structure and Biochemistry of Fe-S Proteins
ISCs occur mainly in a rhombic arrangement [2Fe-2S] or a cubane form [4Fe-4S]. The [2Fe-2S] rhombic cluster consists of two bridging sulfide ions coordinating two iron ions to four cysteines (or two cysteines and two histidines) in a protein (Fig. 1b). Recent reports indicate an unusual occurrence involving three cysteine residues and one histidine in the coordination of a [2Fe-2S] cluster [24]. The [2Fe-2S] clusters occur in two oxidation states, oxidized (both irons are +3) and reduced (one iron is +2 and one is +3) [25]. In the cubane structure, four iron atoms and four inorganic sulfur atoms are coordinated to four sulfhydryl side chains of cysteines (Fig. 1d) [26]. Clusters of this kind are subdivided into two categories, low- and high-potential types. Low-potential [4Fe-4S] clusters switch between oxidized and reduced states of [2Fe3+, 2Fe2+] and [Fe3+, 3Fe2+], respectively. The high-potential subgroup shuttles between an oxidized state of [3Fe3+, Fe2+] and a reduced [2Fe3+, 2Fe2+] [5, 27].
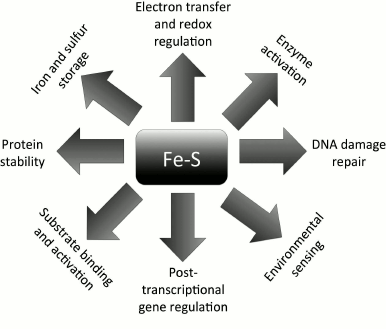
ISCs serve a wide-range of functions (Fig. 2), and their roles can be categorized according to the chemistry involved in their reactions. Oxidation-reduction reactions and electron transfer represent the main function of ISCs [28] and they serve a major role in cellular redox regulation and homeostasis. Because they have the ability to reversibly bind iron and sulfur, ISCs can also be used for Fe and S storage in the activation of specific enzymes and/or substrates (e.g. the aconitase reaction in the citric acid cycle [10, 19]). Interestingly, ISCs are also involved both directly and indirectly in regulation of gene expression [29, 30] and DNA replication [31, 32] and repair [12, 33]. Moreover, they can also enhance protein stability in vitro and in vivo [32, 34, 35].
Fig. 2. Functional versatility of Fe-S clusters. ISCs in proteins serve multifaceted and unrelated functions. They are involved in redox sensing, DNA metabolism and gene activation, iron and sulfur storage, protein stability, and environmental sensing.
Assembly (the ISC Machinery)
We consider here only the ISC machinery in mitochondria. Paul and Lill provide a review of the biogenesis of cytosolic and nuclear Fe-S proteins via the CIA pathway [23]. The Fe-S biogenesis pathway was first elucidated in bacteria and has been shown to be highly conserved in all species including mammals [19, 36]. Due to the complexity of and requirements for Fe-S proteins in various cellular compartments in eukaryotes, the pathway is more elaborate and requires export proteins for the crosstalk between compartments. In yeast, assembly of ISCs occurs mainly in mitochondria, whereas in mammalian cells it also occurs in the cytoplasm and nucleus [37]. Regardless of the organism, the ISC system requires a source of electrons, iron, and sulfur atoms from cysteine.
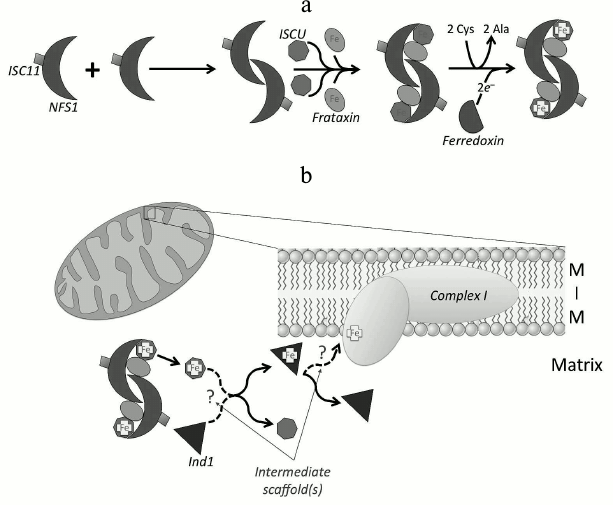
In mammalian mitochondria, Fe-S biogenesis (Fig. 3a) begins with dimerization of a cysteine desulfurase (NFS1) to form a complex with two molecules of the scaffolding protein ISCU at each end of the dimer [38]. NFS1 is stable only when bound to partner protein ISD11 [7, 39], and it is bound in the mitochondrial matrix, cytoplasm, and nucleus [8]. Both ISCU subunits bind inorganic sulfur that is provided by NFS1 in the conversion of two cysteine residues into alanine. The sulfur is then coordinated with iron that is already bound covalently by ISCU [22, 40, 41] via linkage to cysteine residues. The source of iron is likely from the protein frataxin [13]. Frataxin has been shown to catalyze the sulfur transfer step that is rate limiting in the synthesis of [2Fe-2S] clusters [42, 43]. Frataxin binds in a groove between NFS1 and ISCU [19] in the preformed complex rather than to its individual components [44], and it induces a conformational change that activates the complex allosterically by accelerating the sulfur transfer reaction [14, 45, 46]. An electron source is needed to finalize the configuration of the nascent Fe-S structure. Some evidence indicates that ferredoxin can donate the required electrons [47]. In vitro, ferredoxin provides the electrons required to couple two [2Fe-2S] clusters to form a [4Fe-4S] on the ISCU scaffold protein [48]. Defects in any part of this pathway can lead to genetic diseases such as leukodystrophy and neuroregression [49], thus highlighting the important role ISCs play in mitochondrial and cellular health.
Fig. 3. Fe-S proteins are synthesized on the mitochondrial ISC machinery. a) The cysteine desulfurase protein (NFS1) in complex with the stabilizing protein ISC11 dimerizes, allowing two molecules of ISCU to bind. The binding of ISCU to both ends of the dimer creates two grooves where two frataxin molecules bind. The functional cysteine desulfurase complex extracts sulfur from cysteine, converting it to alanine. Frataxin supplies the iron, and ferredoxin donates electrons to form the ISC on ISCU. b) Ind1 is an ISC-targeting factor that has a role in the assembly of the N modules in complex I (cI) in mitochondria. It may acquire the ISC directly from ISCU, or more likely, by an indirect route involving other intermediate scaffolds. Ind1 may be targeted to the mitochondrial membrane either before or after ISC insertion, where it is involved in the transfer of [4Fe-4S] clusters to cI, either directly or again perhaps via other intermediate partners (dashed arrows and question marks) [54, 151].
After ISC incorporation on the core complex, it can then be transferred to target proteins using co-chaperones such as mammalian mitochondrial HSC20 [50] (or bacterial HscB [51]). HSC20 interacts with a protein (containing LYR motif) that is a target ISC protein or an assembly protein for the ISC protein, forming a complex that consists of chaperone–co-chaperone–ISCU–apo-ISC protein (HSPA9–HSC20–ISCU–apo-ISC protein) [52]. Many Fe-S proteins and Fe-S protein assembly subunits in respiratory complexes II and III (cII and cIII) acquire ISCs in this manner [52, 53]. Alternatively, some of the ISCs in complex I (cI) are delivered via a different route. The mitochondrial P-loop NTPase Ind1 (an ISC assembly protein known to be required specifically for NADH dehydrogenase) transfers ISCs to apoproteins at the terminal stage in the ISC assembly process in mitochondria [54-56]. In humans, it is also called NUBPL (nucleotide-binding protein-like) [55]. Ind1 binds transiently a [4Fe-4S] cluster and transfers it to cI [54, 55, 57] (Fig. 3b). Interestingly, Ind1 shows strong specificity for cI proteins in yeasts and humans [54, 55]. The Ind1 deletion mutant in the yeast Yarrowia lipolytica shows only ~30% residual activity and ~20% of the relative abundance of cI compared to wild type, suggesting that the decreased activity is caused by a decrease in the cI level [54]. Likewise, a knockdown mutant of Ind1 in human HeLa cells showed a 3- to 4-fold decrease in cI activity and reduced cI assembly [55]. However, no such reduction in activity or assembly was detected in other mitochondrial Fe-S proteins such as aconitase, cII or cIII, all of which contain ISCs as cofactors [54, 55]. Despite its strong specificity for cI, the 30% residual activity of cI in the Ind1 deletion mutant suggests the possibility of involvement of other ISC delivery proteins for insertion of ISC to cI in addition to Ind1.
Ind1 may also serve other roles in mitochondria. In Drosophila melanogaster mitochondria, a physical interaction between a homolog of Ind1 (CG3262) and the mitochondrial DNA (mtDNA) replicative helicase was found by high-throughput co-affinity purification coupled with mass spectrometry [58], suggesting an expanded role for Drosophila Ind1 in mitochondrial DNA replication. This notion is supported by our recent discovery that the Drosophila mtDNA replicative helicase contains an ISC [34] (see section “Helicases”).
Fe-S CLUSTERS IN MITOCHONDRIAL RESPIRATION
Mitochondrial respiration is the main energy-yielding mechanism in aerobic eukaryotes. In oxidative phosphorylation, electrons donated by NADH and FADH2 are transferred to the last electron acceptor, oxygen, passing through redox centers in four protein complexes in the mitochondrial inner membrane. Many respiratory complex proteins coordinate ISCs that are essential for their activity, and mutations in genes encoding proteins required for biogenesis of Fe-S proteins result in reduced activity of the respiratory chain [59, 60]. Electrons can be transferred directly by the reduction of Fe3+ to Fe2+ in cytochromes and Fe-S proteins. Unlike other redox centers like flavins, quinones, and other metals, both hemes and ISCs are likely to form a chain [61]. Respiratory cI (NADH:ubiquinone oxidoreductase) and cII (succinate:ubiquinone oxidoreductase) contain multiple ISCs and transfer electrons one at a time to ubiquinone (Q) by establishing electron tunneling chains. Complex III contains only a single Fe-S protein.
Complex I (NADH:Ubiquinone Oxidoreductase)
Electrons from NADH enter the respiratory chain through the cI gateway. Complex I is a multisubunit mega protein carrying eight ISCs in five of its fourteen core subunits. The nomenclature of the eight ISCs is based on EPR signals [62, 63]: two (N1a and N1b) are [2Fe-2S] clusters, and six are [4Fe-4S] clusters (N2, N3, N4, N5, N6a, and N6b). Some prokaryotes including Thermus thermophilus and Escherichia coli contain an additional [4Fe-4S] cluster N7 in a subunit that is thought to play a role in assembly and/or structural stability rather than electron transfer due to its distal position from the main redox chain [64]. All the ISCs locate in the peripheral arm of cI, which has a hydrophilic domain and protrudes into the matrix. Ubiquinone positioned at the interface between the peripheral arm and the membrane arm [65, 66] is reduced by the N2 cluster, the terminal cluster in the cI redox chain. Complex I transfers two electrons from NADH to ubiquinone (Q) in an exergonic process that is tightly coupled to the endergonic translocation of four protons across the membrane into the acidic intermembrane space [67]. The ISCs in cI are described below; individual proteins are indicated by their human/bovine designations.
N3: NDUFV1/51 kDa. Cluster N3 is a [4Fe-4S] cluster that is positioned within ~8 Å of flavin mononucleotide (FMN) in subunit NDUFV1/51 kDa, and is the first ISC in the redox chain. One electron from FMN is transferred by N3 to N1b in NDUFS1/75 kDa via intersubunit transfer [68, 69].
N1b, N4, N5: NDUFS1/75 kDa. NDUFS1/75 kDa carries three ISCs; N1b is a [2Fe-2S] cluster and N4 and N5 are [4Fe-4S] clusters [70]. NDUFS1/75 kDa transfers an electron from N1b to N4 to N5. Notably, N5 is coordinated by three cysteines and one histidine, so its EPR properties differ from clusters coordinated by four cysteine residues [70]. The longest edge-to-edge distance between ISCs is that between N5 in NDUFS1/75 kDa and N6a in NDUFS8/TYKY. Thus, the electron transfer rate between N5 and N6a is the rate-limiting step in the electron transfer pathway within the ISC chain in cI. Nonetheless, molecular dynamic simulations suggest that a water molecule in the intersubunit space can enhance the rate of transfer by near three orders of magnitude [71].
N6a, N6b: NDUFS8/TYKT. NDUFS8/TYKT has two [4Fe-4S] clusters and transfers an electron from N6a to N6b. N6a is located near the interface of the N module and the Q module of cI near the zinc-binding site of a cI accessory subunit, NUMM (in Y. lipolytica) or NDUFS6 (in humans) [72]. Mutations in Zn-coordinating residues of NUMM compromise proper assembly of cI, and deletion of NUMM causes reduction in the EPR signal of N6a, suggesting that stable insertion of N6a requires the Zn-binding site in NUMM [72].
N2: NDUFS7/PSST. Cluster N2 is coordinated in NDUFS7 near its interface with NDUFS8. Due to its higher redox midpoint potential, N2 receives electrons from other clusters as an electron sink [70] and transfers them to ubiquinone exiting the electron transport chain at cI [73]. The midpoint potential of N2 shows a pH dependence, becoming more positive at lower pH values [74]. This pH dependence is due to the protonated group, His226 of the 49 kDa (NDUFS2) subunit in the case of Y. lipolytica. Mutation of this histidine abolishes the pH dependence, but surprisingly does not affect the proton pumping mechanism, implying that the ISC redox chain is not linked directly to proton pumping. Rather, it is suggested that the interaction between N2 and semiquinone species may be linked to a redox-driven coupling mechanism [73, 75, 76].
N1a: NDUFV2/24 kDa. NDUFV2/24 kDa carries a [2Fe-2S] cluster, N1a, in a hydrophobic surrounding. N1a is not part of the main electron redox chain; its position is too remote and it has a very low mid-point potential. Because the mid-point potential is higher than that of flavosemiquinone, it is thought that one of two electrons from FMN is transferred to N1a and the other to N3 [63]. It has also been suggested that N1a serves a role in preventing ROS production [77]. Though its specific role has not been elucidated, N1a is conserved across species, suggesting its importance.
Complex II (Succinate:Ubiquinone Oxidoreductase; Succinate Dehydrogenase (SDH))
Complex II comprises four nuclear-encoded polypeptides; a flavoprotein (SDHA) and a Fe-S protein (SDHB) form a hydrophilic head in the matrix and are tethered to a membrane anchor domain that consists of SDHC and SDHD [78]. Complex II transfers electrons derived from the oxidation of succinate to fumarate in the citric acid cycle via FADH2 to ultimately reduce the mobile electron carrier ubiquinone, coupling the citric acid cycle and the electron transport chain (ETC). Complex II (SDH) is the only component of the citric acid cycle that is membrane-bound. It is distinguished from other complexes in the ETC by its inability to pump protons, in addition to its lack of any subunit that is encoded by mitochondrial DNA. The two reactions catalyzed by cII, the oxidation of succinate in the flavoprotein (SDHA) and the reduction of ubiquinone in the membrane-anchored domain (SDHC + SDHD), are linked via electron transport through three ISCs in SDHB [79].
Succinate dehydrogenase B (SDHB). SDHB contains three different ISCs: [2Fe-2S], [4Fe-4S], and [3Fe-4S]. The rhombic [2Fe-2S] cluster is ligated by four cysteine residues in the N-terminal domain comprising an α-helix and five β-strands. It is located adjacent to FAD in the flavoprotein (SDHA). Both [4Fe-4S] and [3Fe-4S] are in the C-terminal domain, which contains six α-helices that interact largely with the membrane anchor domain. The three ISCs are aligned almost linearly. Each edge-to-edge distance is less than 14 Å, indicating a favorable electron transfer [61]. The [3Fe-4S] cluster is in the ISC chain in SDHB and lies 7.1 Å away from ubiquinone and 13.3 Å from heme [61, 78]. Thus, reduction of ubiquinone occurs prior to that of hemes, as is anticipated from their respective redox potentials [78].
Insertion of the ISCs into apo-SDHB occurs in the mitochondrial matrix prior to formation of a heterodimer with SDHA; it is guided by HSC20, a co-chaperone in the ISC biogenesis pathway [80]. Rouault and coworkers [52, 53] showed in a yeast two-hybrid screen that SDHB has three independent binding sites for HSC20; two sites have a L(I)YR motif and one has a KKx6-10KK motif. These are the two most prevalent consensus sequences for HSC20 binding and are found in Fe-S cluster recipient proteins [52, 53].
SDHAF1, an assembly protein for cII, also contains a L(I)YR motif, implying that it may enable the insertion of ISCs into SDHB, using its interaction with HSC20 [52]. Co-immunoprecipitation analysis and mitochondrial subfractionation coupled with Blue Native Polyacrylamide Gel Electrophoresis (BN-PAGE) results show that three clusters are transferred by a chaperone/co-chaperone system through either the HSC20–HSPA9–ISCU–SDHB complex or the HSC20–HSPA9–ISCU–SDHB–SDHAF1 complex [52]. As expected, mutations in SDHAF1 can cause succinate dehydrogenase deficiency manifesting as infantile leukoencephalopathy with accumulation of blood succinate and lactate [21].
Complex III (Ubiquinol:Cytochrome c-Oxidoreductase)
Electrons are shuttled from cI and/or cII via ubiquinol (the reduced form of ubiquinone) to cIII. Complex III transfers an electron from ubiquinol to cytochrome c and recycles the other electron from ubiquinol for proton motive force generation through the Q-cycle mechanism in a bifurcated fashion. Structural studies demonstrate that cIII is a dimer [81-83] that contains three essential redox subunits: cytochrome b, cytochrome c1, and the Fe-S protein (ISP), although the total number of subunits varies from three in prokaryotes to eleven in humans [84]. It harbors three types of redox centers: two b-type hemes, one c-type heme, and a [2Fe-2S] cluster. ISP mediates one electron transfer from ubiquinol to cytochrome c1 modulated by another electron transfer pathway through cytochrome b [84-86].
Iron sulfur protein (ISP). ISP in cIII is also called “Rieske” protein because it has a Rieske-type [2Fe-2S] cluster that is coordinated by two cysteines and two histidines [82, 85-88]. ISP is anchored to the mitochondrial inner membrane by a transmembrane helix with a soluble extramembrane domain called the extrinsic domain at its C-terminus [81, 82, 89]. The extrinsic domain (ISP-ED) harbors the [2Fe-2S] cluster in the intermembrane space. This domain moves to the Qo site or to cytochrome c1 depending on the occupancy of the Qo site by inhibitors, enabling the [2Fe-2S] cluster to accept an electron from the site of oxidation of ubiquinol and to donate the electron to an oxidized cytochrome c1, respectively [81, 82, 90]. Although it is clear that a substantial conformational change of the ISP-ED is necessary for the bifurcated electron transfer to overcome the >14 Å distance and unfavorable rate, the detailed mechanism is still unknown. Two models have been proposed to explain movement: (i) the binding affinity modulated ISP-ED motion switch model [84, 90] and (ii) the two-position model [91]. Interestingly, a mutation in cytochrome b of Rhodobacter capsulatus (G167P) shifts the movement of ISP-ED toward positions far from the Qo site and induces the production of superoxide radicals [92]. A corresponding mutation in the human enzyme (S151P) has been identified as a mitochondrial disease-related mutation [92]. Gurung et al. reported that cIII lacking its ISC creates a proton leak, suggesting a role for the [2Fe-2S] cluster in gating a proton channel [86].
Insertion of the [2Fe-2S] cluster in ISP is assisted by LYRM7, an assembly factor shown to bind to it in human cIII [93]. LYRM7 is co-immunoprecipitated with HSC20. That the level of ISP is reduced after knockdown of HSC20 suggests LYRM7 also participates with the HSC20–HSPA9–ISCU complex for ISP assembly in cIII, as does SDHAF in assembly of cII [52].
Fe-S CLUSTERS IN MITOCHONDRIAL METABOLISM
Fe-S clusters play important roles in the various metabolic pathways within mitochondria. We discuss below the recent literature on Fe-S involvement in mitochondrial metabolism.
Citric Acid Cycle
The citric acid cycle is the central cyclic pathway by which the carbon skeleton of glucose is released as carbon dioxide, producing energy in the form of ATP and reduced coenzymes [94]. A molecule of oxaloacetate is regenerated in a sequence of eight enzyme-catalyzed reactions. Three enzymes contain ISCs: aconitase (which can be considered as a moonlighting enzyme with dual roles involving its ISC), succinate dehydrogenase (common to both the citric acid cycle and the electron transport chain, see section “Fe-S Clusters in Mitochondrial Respiration”), and fumarase (in several bacteria and a single archaeon, to date).
Aconitase. Aconitase, the second enzyme of the citric acid cycle, catalyzes the conversion of citrate to isocitrate via an alkene intermediate (cis-aconitate). It has two conformations depending on its activity, both of which coordinate an ISC. The inactive protein comprises four domains and binds a [3Fe-4S] cluster [95]. Once activated, it acquires another iron atom to form a [4Fe-4S] center [96, 97]. The cluster is coordinated by three cysteine residues, and upon insertion, its labile iron is coordinated by water molecules. The additional iron atom is crucial for the activity of the enzyme and can be removed by a variety of oxidants. For example, thiocyanate (an oxidant elevated in the arteries of smokers) releases it both in the isolated enzyme and in cultured cells, leading to protein dysfunction [98]. Chromate ions also cause the oxidative inactivation of aconitase and other Fe-S proteins [99]. Similarly, low doses of the strong oxidant peroxynitrite release the labile iron [100].
A cytosolic isozyme of mitochondrial aconitase termed iron-regulatory protein (IRP) or iron-responsive element binding protein (IRE-BP) [101] has aconitase activity and serves another role in cellular iron homeostasis, a function that is also mediated through its ISC [102]. It is an mRNA-binding protein that binds iron responsive elements (IRE) in mRNA, resulting in either its stabilization or degradation, thus providing a posttranscriptional control point. The key players in cellular iron homeostasis are ferritin (an iron-storage protein) and the transferrin receptor (a membrane gateway for the cellular entry of iron from blood). Apo-aconitase (carrying a [3Fe-4S] cluster because of low cytosolic iron concentration) binds and stabilizes the mRNA for the transferrin receptor, promoting protein production and increasing iron transport into the cytoplasm. At the same time, apo-aconitase binds to the ferritin mRNA and prevents its translation under cellular conditions of low iron [94]. When iron levels are elevated in the cytoplasm, apo-aconitase acquires the labile iron and induces a conformational change to holo-aconitase carrying a [4Fe-4S] cluster, leading to the availability of free ferritin mRNA and subsequent ferritin production [103]. As with the mitochondrial isoform, cytosolic aconitase is also regulated through its ISC [104]. Aconitase activity is inhibited in cells treated with thiocyanate resulting in increased IRP-1 activity and ultimately higher levels of iron in the cytoplasm, possibly resulting in toxic effects [98]. Interestingly, mitochondrial aconitase in yeast has also been shown to serve a role in mitochondrial DNA (mtDNA) maintenance independent of its catalytic activity [105]. Whether mitochondrial or cytosolic, aconitase presents a case of dual function mediated by its ISC.
DNA Metabolism
Until recently, knowledge of the presence of Fe-S clusters in enzymes specialized in nucleic acid metabolism was rare [106]. Now numerous enzymes involved in DNA and RNA transactions have been shown to carry one or more ISCs of various types and structures. The presence of ISCs in primases, helicases, nucleases, ligases, glycosylases, polymerases, and transcription factors has proved to be essential for protein structure and function [107]. To date, only a few proteins in mtDNA metabolism have been demonstrated to contain ISCs, but this number is highly likely to become substantially larger. ISCs function as a typical redox-sensing, electron-transferring cofactor in some nucleic acid processing proteins. For example, the EndoIII and MutY DNA repair glycosylases sense DNA damage via charge transfer within their Fe-S clusters [108, 109]. Some RNA metabolism enzymes use ISCs to transfer electrons to S-adenosylmethionine to mediate methylation of target rRNAs and tRNAs [110]. In addition, many transcription factors that sense nitric oxide regulate gene expression via small changes in their ISCs [4]. ISCs also serve multifarious other roles. We present below a short description of current knowledge; the table summarizes recent findings about some ISC-carrying enzymes in DNA metabolism, including the several that have been identified in mitochondria.
Structure and function of ISCs in some DNA metabolism enzymes
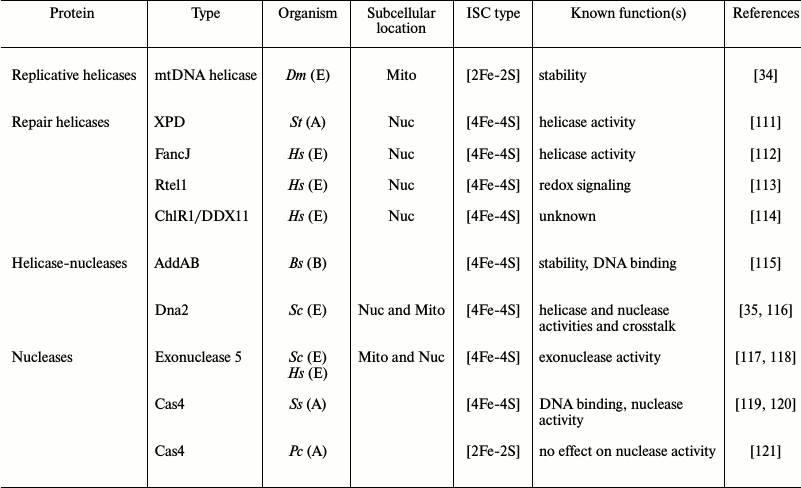
Notes: A, Archaea; B, Bacteria; E, Eukarya; Bs, Bacillus
subtilis; Pc, Pyrobaculum calidifontis; Ss,
Sulfolobus solfataricus; St, Sulfolobus tokodaii;
Sc, Saccharomyces cerevisiae; Dm, Drosophila
melanogaster; Hs, Homo sapiens; Mito, mitochondria;
Nuc, nucleus.
Helicases. DNA helicases play a central role in nucleic acid metabolism to unwind double-stranded DNA using the energy provided by nucleotide hydrolysis to translocate along and provide access to single-stranded DNA in the process of transcription, replication, recombination, and repair [122].
ISCs in helicases have been identified largely in DNA helicase superfamilies 1 and 2 (SF1 and SF2) [123]. An exception is the mitochondrial replicative DNA helicase, which is a SF4 helicase [34, 124]. The XPD/FANCJ family including XPD, FancJ, Chlr1, and RTEL has been studied extensively due to its strong association with human diseases [125]. XPD was the first ISC-carrying DNA repair helicase in SF2 to be identified [126]. Its [4Fe-4S] cluster is located in the core helicase domain. Both structural [127, 128] and biochemical [129] analyses show that the ISC in XPD is proximal to the duplex DNA strand separation site, suggesting a role for the ISC in determination of the unwinding point. ISCs in XPD and FancJ are also essential for helicase activity in nucleotide excision repair [112, 126]. Consequently, mutations in human XPD cause several genetic diseases such as Xeroderma pigmentosum, Cockayne syndrome, Fanconi anemia, and trichothiodystrophy [112, 126]. Furthermore, Fe-S-cluster DNA helicases were shown to be inhibited potently by protein–DNA interactions, thereby affecting DNA metabolism. For example, the helicase activity of FancJ is inhibited by shelterin proteins that bind telomeres [130].
Mitochondrial replicative DNA helicase. The mitochondrial replicative DNA helicase from Drosophila melanogaster is the only known ISC-containing replisome protein identified in mitochondria to date. Our group has shown recently that its N-terminal domain contains a [2Fe-2S] cluster that is essential for protein stability in vitro [34]. Its ISC is bound by the homologous cysteine residues that coordinate zinc in the primase-helicase from bacteriophage T7. This evolutionary switch from zinc to iron binding is intriguing, particularly because it resides in the N-terminal part of the protein that has not yet been ascribed a clear function.
Helicase-nucleases. Nucleases cleave phosphodiester bonds by various catalytic mechanisms [131]. Some nucleases have evolved into bifunctional enzymes carrying two catalytic domains in a single polypeptide: a helicase domain for unwinding DNA and a nuclease domain for phosphodiester bond cleavage [132]. Two helicase-nucleases in the RecB family have been shown to carry ISCs: AddAB and Dna2. AddAB is a well-studied Fe-S helicase-nuclease from the bacterium Bacillus subtilis [133]; it contains a 4Fe-4S cluster in its C-terminal domain with roles in maintaining the structural stability of the nuclease domain and binding to DNA ends [115]. Similar structures and roles have been found in other nucleases outside the helicase-nuclease group, including yeast exonuclease 5 [117] and the archaeal nuclease Cas4 from Sulfolobus solfataricus [119].
Dna2. Dna2, a helicase-nuclease in the RecB family, was identified as an ISC-containing enzyme that localizes to both the nucleus and mitochondria [35, 134, 135]. Its ISC was predicted based on the presence of a conserved ISC signature motif within the nuclease active site [116]. Pokharel and Campbell demonstrated the presence of the ISC in the Dna2 helicase-nuclease from Saccharomyces cerevisiae by UV-vis spectrometry and mutagenesis of the ISC-ligating cysteines, which causes reduction in both its nuclease and ATPase activities, suggesting a possible role of the ISC in linking its nuclease and helicase functions [35]. However, variants lacking the cluster showed no significant difference in DNA binding [35]. Dna2 is involved in maintaining the integrity of both the nuclear and mitochondrial genomes [134, 135]. Its role in yeast nuclear DNA replication and repair in Okazaki fragment processing and double-strand break repair, respectively, are well-characterized [136]. Zheng et al. demonstrated that human Dna2 stimulates mtDNA polymerase activity and is involved in removal of RNA primers and intermediates of long-patch base excision repair in mitochondria [134].
Nucleases. The structural signature of the nuclease domain in which the ISC is bound in the AddAB helicase-nuclease is also found in other nucleases including exonuclease 5 and Cas4 [107, 115]. An in vitro study showed that the ISC of the Cas4 nuclease from S. solfataricus serves a role in protein stability [119]. The crystal structure of Cas4 revealed a toroidal form comprising five dimers, with each protomer containing a 4Fe-4S cluster that is required for DNA binding and cleavage activity [120]. Interestingly, Cas4 from Pyrobaculum calidifontis carries a 2Fe-2S cluster, indicating species-specific differences. However, the loss of the ISC apparently has no effect on nuclease activity [121].
Yeast exonuclease 5. Yeast exonuclease 5 (EXO5) localizes to mitochondria and serves an essential role in mitochondrial genome stability [117]. The presence of an ISC was predicted by sequence alignment with RecB family nucleases, and it shares conserved cysteine residues with those in the nuclease domain of AddAB [117]. The presence of an ISC was subsequently demonstrated by UV-vis spectrometry in its human homolog, hEXO5 [118]. A mutant lacking the ISC in hEXO5 exhibits only ~10-20% residual nuclease activity; however, it may function only in nuclear DNA repair because unlike yeast EXO5, hEXO5 gene lacks a mitochondrial leader sequence [118].
Apoptosis
Mitochondria are not only essential for the vitality of cells; they serve a strategic role in committing cells to undergo programmed cell death [137]. Intrinsic apoptosis is mediated by the intactness of the mitochondrial outer membrane. A breach in the outer membrane results in the release of proteins from the intermembrane space (specifically cytochrome c), and a cascade of events eventually leads to apoptotic cell death [138, 139]. Fe-S proteins play multifarious roles in mitochondrial iron homeostasis and apoptosis. MitoNEET is an integral mitochondrial outer membrane protein that has been implicated in iron homeostasis in both mitochondria and the cytoplasm [11, 140, 141]. It contains a redox-active [2Fe-2S] cluster [24, 142]. The induction of cell death through treatment with TNFα is mediated by the binding of the Stat3–Grim–19 complex to mitoNEET, forcing the rapid release of its ISC. Mitochondria then accumulate iron in the matrix, inducing the formation of reactive oxygen species and mitochondrial injury accompanied by cell death [143]. MitoNEET has been shown recently to act as an ISC transfer protein [144] that uses a redox switch mechanism to regulate transfer [145]; its redox-sensing role helps to combat oxidative injury by recovery of labile Fe-S proteins [146]. In pancreatic β cells, overexpression of mitoNEET leads to an increased sensitivity to TNFα cytotoxicity, and the effects of TNFα can be attenuated if it is unable to transfer its ISC (via pharmacological intervention) [147]. MitoMEET has also been implicated in the proliferation of human breast cancer cells and promotion of tumor growth [148].
Cancer can also be induced by the absence of p53, the “guardian of the genome”. Lack of p53 in tumor cells was shown recently to decrease utilization of mitochondrial iron by downregulation of the expression of human frataxin [149]. Cellular iron deficiency is also detrimental for lipid synthesis and will eventually alter the properties and functions of biological membranes [150], thereby affecting multiple mitochondrial pathways.
The CIA machinery for ISC assembly in the cytoplasm is also involved in mitochondrial integrity and/or apoptosis. Human cytokine induced apoptosis inhibitor (CIAPIN1) belongs to the anamorsin protein family and has been shown to function as an antiapoptotic protein through the regulation of Bcl-2 and Bax [151]. Anamorsin carries a [2Fe-2S] cluster and functions early in the CIA pathway as part of an electron transfer chain [152]. The yeast homolog of anamorsin, Dre2, is also a Fe-S protein to which electrons are transferred from Tah18 in the early steps of CIA [153]. The interaction of Tah18 and Dre2 is lost when yeast cells are exposed to lethal doses of H2O2 and Tah18 is relocalized to mitochondria. There it mediates the loss of outer membrane integrity, promoting apoptosis. Interestingly, anamorsin was able to substitute for yeast Dre2 and in its interaction with Tah18 [154].
CONCLUSIONS AND FUTURE DIRECTIONS
The requirements for and versatility of iron-sulfur clusters in myriad metabolic pathways in organisms across taxa has led to intensive research on their structural and functional properties. The roles of ISCs in various cellular compartments and the influence of environmental factors upon them have been explored in detail in multiple systems. In mitochondria, their function in respiration is well documented, as is the pathway of their synthesis and transfer to recipient proteins. Other areas of mitochondrial Fe-S biology are less well explored, and the function of ISCs in mitochondrial nucleic acid metabolism remains somewhat enigmatic. To our knowledge, only three such proteins have been identified to date, each playing an important role in DNA replication and/or repair, and perhaps recombination, but their evolutionary histories are not readily apparent; each of them may not be present in all animal taxa and/or they may not all carry an ISC. For example, the mtDNA replicative helicase in insect lineages is likely to carry a [2Fe-2S] cluster, which is likely not present in mammals (or perhaps any vertebrate species), and this enzyme is absent in yeast, despite its evolutionary relatedness to the primase-helicase of bacteriophage T7. To date, no ISC-containing protein in mitochondrial RNA metabolism has been identified, and given the structural and catalytic properties of proteins involved in mitochondrial transcription, RNA processing and storage, and translation, this is surprising. Thus, there remains much to explore in the realm of ISC biology within the mitochondrion.
Acknowledgements
We thank Dr. Grzegorz Ciesielski for critical reading of the manuscript.
Laurie S. Kaguni acknowledges research support from the National Institutes of Health (project No. GM45295). Johnny Stiban acknowledges travel support provided by the Welfare association in conjunction with the Bank of Palestine, administered via Zamalah scholarship.
REFERENCES
1.Baker, H. M., Anderson, B. F., and Baker, E. N.
(2003) Dealing with iron: common structural principles in proteins that
transport iron and heme, Proc. Natl. Acad. Sci. USA, 100,
3579-3583.
2.Beinert, H., Holm, R. H., and Munck, E. (1997)
Iron-sulfur clusters: nature’s modular, multipurpose structures,
Science, 277, 653-659.
3.Beinert, H. (2000) Iron-sulfur proteins: ancient
structures, still full of surprises, J. Biol. Inorg. Chem.,
5, 2-15.
4.Kiley, P. J., and Beinert, H. (2003) The role of
Fe-S proteins in sensing and regulation in bacteria, Curr. Opin.
Microbiol., 6, 181-185.
5.Johnson, D. C., Dean, D. R., Smith, A. D., and
Johnson, M. K. (2005) Structure, function, and formation of biological
iron-sulfur clusters, Annu. Rev. Biochem., 74,
247-281.
6.Brzoska, K., Meczynska, S., and Kruszewski, M.
(2006) Iron-sulfur cluster proteins: electron transfer and beyond,
Acta Biochim. Pol., 53, 685-691.
7.Wiedemann, N., Urzica, E., Guiard, B., Muller, H.,
Lohaus, C., Meyer, H. E., Ryan, M. T., Meisinger, C., Muhlenhoff, U.,
Lill, R., and Pfanner, N. (2006) Essential role of Isd11 in
mitochondrial iron-sulfur cluster synthesis on Isu scaffold proteins,
EMBO J., 25, 184-195.
8.Shi, Y., Ghosh, M. C., Tong, W. H., and Rouault, T.
A. (2009) Human ISD11 is essential for both iron-sulfur cluster
assembly and maintenance of normal cellular iron homeostasis, Hum.
Mol. Genet., 18, 3014-3025.
9.Tong, W. H., and Rouault, T. (2000) Distinct
iron-sulfur cluster assembly complexes exist in the cytosol and
mitochondria of human cells, EMBO J., 19,
5692-5700.
10.Tong, W. H., and Rouault, T. A. (2006) Functions
of mitochondrial ISCU and cytosolic ISCU in mammalian iron-sulfur
cluster biogenesis and iron homeostasis, Cell Metab., 3,
199-210.
11.Tamir, S., Paddock, M. L., Darash-Yahana-Baram,
M., Holt, S. H., Sohn, Y. S., Agranat, L., Michaeli, D., Stofleth, J.
T., Lipper, C. H., Morcos, F., Cabantchik, I. Z., Onuchic, J. N.,
Jennings, P. A., Mittler, R., and Nechushtai, R. (2015)
Structure-function analysis of NEET proteins uncovers their role as key
regulators of iron and ROS homeostasis in health and disease,
Biochim. Biophys. Acta, 1853, 1294-1315.
12.Fuss, J. O., Tsai, C. L., Ishida, J. P., and
Tainer, J. A. (2015) Emerging critical roles of Fe-S clusters in DNA
replication and repair, Biochim. Biophys. Acta, 1853,
1253-1271.
13.Stemmler, T. L., Lesuisse, E., Pain, D., and
Dancis, A. (2010) Frataxin and mitochondrial Fe-S cluster biogenesis,
J. Biol. Chem., 285, 26737-26743.
14.Bridwell-Rabb, J., Fox, N. G., Tsai, C. L., Winn,
A. M., and Barondeau, D. P. (2014) Human frataxin activates Fe-S
cluster biosynthesis by facilitating sulfur transfer chemistry,
Biochemistry, 53, 4904-4913.
15.Rouault, T. A., and Tong, W. H. (2005)
Iron-sulphur cluster biogenesis and mitochondrial iron homeostasis,
Nat. Rev. Mol. Cell Biol., 6, 345-351.
16.Lill, R., and Muhlenhoff, U. (2006) Iron-sulfur
protein biogenesis in eukaryotes: components and mechanisms, Annu.
Rev. Cell Dev. Biol., 22, 457-486.
17.Lill, R., Dutkiewicz, R., Elsasser, H. P.,
Hausmann, A., Netz, D. J., Pierik, A. J., Stehling, O., Urzica, E., and
Muhlenhoff, U. (2006) Mechanisms of iron-sulfur protein maturation in
mitochondria, cytosol and nucleus of eukaryotes, Biochim. Biophys.
Acta, 1763, 652-667.
18.Napier, I., Ponka, P., and Richardson, D. R.
(2005) Iron trafficking in the mitochondrion: novel pathways revealed
by disease, Blood, 105, 1867-1874.
19.Rouault, T. A. (2012) Biogenesis of iron-sulfur
clusters in mammalian cells: new insights and relevance to human
disease, Dis. Models Mech., 5, 155-164.
20.Beilschmidt, L. K., and Puccio, H. M. (2014)
Mammalian Fe-S cluster biogenesis and its implication in disease,
Biochimie, 100, 48-60.
21.Maio, N., Ghezzi, D., Verrigni, D., Rizza, T.,
Bertini, E., Martinelli, D., Zeviani, M., Singh, A., Carrozzo, R., and
Rouault, T. A. (2015) Disease-causing SDHAF1 mutations impair transfer
of Fe-S clusters to SDHB, Cell Metab., 23, 292-302.
22.Lill, R. (2009) Function and biogenesis of
iron-sulphur proteins, Nature, 460, 831-838.
23.Paul, V. D., and Lill, R. (2015) Biogenesis of
cytosolic and nuclear iron-sulfur proteins and their role in genome
stability, Biochim. Biophys. Acta, 1853, 1528-1539.
24.Wiley, S. E., Paddock, M. L., Abresch, E. C.,
Gross, L., Van der Geer, P., Nechushtai, R., Murphy, A. N., Jennings,
P. A., and Dixon, J. E. (2007) The outer mitochondrial membrane protein
mitoNEET contains a novel redox-active 2Fe-2S cluster, J. Biol.
Chem., 282, 23745-23749.
25.Leggate, E. J., Bill, E., Essigke, T., Ullmann,
G. M., and Hirst, J. (2004) Formation and characterization of an
all-ferrous Rieske cluster and stabilization of the [2Fe-2S](0) core by
protonation, Proc. Natl. Acad. Sci. USA, 101,
10913-10918.
26.Meyer, J. (2008) Iron-sulfur protein folds,
iron-sulfur chemistry, and evolution, J. Biol. Inorg. Chem.,
13, 157-170.
27.Johnson, M. K., and Smith, A. D. (2005)
Iron-sulfur proteins, in Encyclopedia of Inorganic Chemistry
(King, R. B., ed.) 2nd Edn., John Wiley & Sons, Chichester, pp.
2589-2619.
28.Ren, B., Duan, X., and Ding, H. (2009) Redox
control of the DNA damage-inducible protein DinG helicase activity via
its iron-sulfur cluster, J. Biol. Chem., 284,
4829-4835.
29.Ding, H., Hidalgo, E., and Demple, B. (1996) The
redox state of the [2Fe-2S] clusters in SoxR protein regulates its
activity as a transcription factor, J. Biol. Chem., 271,
33173-33175.
30.Ramon-Garcia, S., Ng, C., Jensen, P. R., Dosanjh,
M., Burian, J., Morris, R. P., Folcher, M., Eltis, L. D., Grzesiek, S.,
Nguyen, L., and Thompson, C. J. (2013) WhiB7, an Fe-S-dependent
transcription factor that activates species-specific repertoires of
drug resistance determinants in actinobacteria, J. Biol. Chem.,
288, 34514-34528.
31.Jain, R., Vanamee, E. S., Dzikovski, B. G., Buku,
A., Johnson, R. E., Prakash, L., Prakash, S., and Aggarwal, A. K.
(2014) An iron-sulfur cluster in the polymerase domain of yeast DNA
polymerase epsilon, J. Mol. Biol., 426, 301-308.
32.Netz, D. J., Stith, C. M., Stumpfig, M., Kopf,
G., Vogel, D., Genau, H. M., Stodola, J. L., Lill, R., Burgers, P. M.,
and Pierik, A. J. (2012) Eukaryotic DNA polymerases require an
iron-sulfur cluster for the formation of active complexes, Nat.
Chem. Biol., 8, 125-132.
33.Zhang, C. (2014) Essential functions of
iron-requiring proteins in DNA replication, repair and cell cycle
control, Protein Cell, 5, 750-760.
34.Stiban, J., Farnum, G. A., Hovde, S. L., and
Kaguni, L. S. (2014) The N-terminal domain of the Drosophila
mitochondrial replicative DNA helicase contains an iron-sulfur cluster
and binds DNA, J. Biol. Chem., 289, 24032-24042.
35.Pokharel, S., and Campbell, J. L. (2012) Cross
talk between the nuclease and helicase activities of Dna2: role of an
essential iron-sulfur cluster domain, Nucleic Acids Res.,
40, 7821-7830.
36.Frazzon, J., and Dean, D. R. (2003) Formation of
iron-sulfur clusters in bacteria: an emerging field in bioinorganic
chemistry, Curr. Opin. Chem. Biol., 7, 166-173.
37.Biederbick, A., Stehling, O., Rosser, R.,
Niggemeyer, B., Nakai, Y., Elsasser, H. P., and Lill, R. (2006) Role of
human mitochondrial Nfs1 in cytosolic iron-sulfur protein biogenesis
and iron regulation, Mol. Cell. Biol., 26, 5675-5687.
38.Shi, R., Proteau, A., Villarroya, M., Moukadiri,
I., Zhang, L., Trempe, J. F., Matte, A., Armengod, M. E., and Cygler,
M. (2010) Structural basis for Fe-S cluster assembly and tRNA
thiolation mediated by IscS protein–protein interactions, PLoS
Biol., 8, e1000354.
39.Adam, A. C., Bornhovd, C., Prokisch, H., Neupert,
W., and Hell, K. (2006) The Nfs1 interacting protein Isd11 has an
essential role in Fe/S cluster biogenesis in mitochondria, EMBO
J., 25, 174-183.
40.Bandyopadhyay, S., Chandramouli, K., and Johnson,
M. K. (2008) Iron-sulfur cluster biosynthesis, Biochem. Soc.
Trans., 36, 1112-1119.
41.Raulfs, E. C., O’Carroll, I. P., Dos
Santos, P. C., Unciuleac, M. C., and Dean, D. R. (2008) In vivo
iron-sulfur cluster formation, Proc. Natl. Acad. Sci. USA,
105, 8591-8596.
42.Fox, N. G., Das, D., Chakrabarti, M., Lindahl, P.
A., and Barondeau, D. P. (2015) Frataxin accelerates [2Fe-2S] cluster
formation on the human Fe–S assembly complex,
Biochemistry, 54, 3880-3889.
43.Fox, N. G., Chakrabarti, M., McCormick, S. P.,
Lindahl, P. A., and Barondeau, D. P. (2015) The human iron–sulfur
assembly complex catalyzes the synthesis of [2Fe-2S] clusters on ISCU2
that can be transferred to acceptor molecules, Biochemistry,
54, 3871-3879.
44.Schmucker, S., Martelli, A., Colin, F., Page, A.,
Wattenhofer-Donze, M., Reutenauer, L., and Puccio, H. (2011) Mammalian
frataxin: an essential function for cellular viability through an
interaction with a preformed ISCU/NFS1/ISD11 iron–sulfur assembly
complex, PLoS One, 6, e16199.
45.Tsai, C. L., and Barondeau, D. P. (2010) Human
frataxin is an allosteric switch that activates the Fe-S cluster
biosynthetic complex, Biochemistry, 49, 9132-9139.
46.Bridwell-Rabb, J., Winn, A. M., and Barondeau, D.
P. (2011) Structure-function analysis of Friedreich’s ataxia
mutants reveals determinants of frataxin binding and activation of the
Fe–S assembly complex, Biochemistry, 50,
7265-7274.
47.Shi, Y., Ghosh, M., Kovtunovych, G., Crooks, D.
R., and Rouault, T. A. (2012) Both human ferredoxins 1 and 2 and
ferredoxin reductase are important for iron-sulfur cluster biogenesis,
Biochim. Biophys. Acta, 1823, 484-492.
48.Chandramouli, K., Unciuleac, M. C., Naik, S.,
Dean, D. R., Huynh, B. H., and Johnson, M. K. (2007) Formation and
properties of [4Fe-4S] clusters on the IscU scaffold protein,
Biochemistry, 46, 6804-6811.
49.Al-Hassnan, Z. N., Al-Dosary, M., Alfadhel, M.,
Faqeih, E. A., Alsagob, M., Kenana, R., Almass, R., Al-Harazi, O. S.,
Al-Hindi, H., Malibari, O. I., Almutari, F. B., Tulbah, S., Alhadeq,
F., Al-Sheddi, T., Alamro, R., AlAsmari, A., Almuntashri, M.,
Alshaalan, H., Al-Mohanna, F. A., Colak, D., and Kaya, N. (2015) ISCA2
mutation causes infantile neurodegenerative mitochondrial disorder,
J. Med. Genet., 52, 186-194.
50.Uhrigshardt, H., Singh, A., Kovtunovych, G.,
Ghosh, M., and Rouault, T. A. (2010) Characterization of the human
HSC20, an unusual DnaJ type III protein, involved in iron-sulfur
cluster biogenesis, Hum. Mol. Genet., 19, 3816-3834.
51.Vickery, L. E., and Cupp-Vickery, J. R. (2007)
Molecular chaperones HscA/Ssq1 and HscB/Jac1 and their roles in
iron-sulfur protein maturation, Crit. Rev. Biochem. Mol. Biol.,
42, 95-111.
52.Maio, N., Singh, A., Uhrigshardt, H., Saxena, N.,
Tong, W. H., and Rouault, T. A. (2014) Cochaperone binding to LYR
motifs confers specificity of iron sulfur cluster delivery, Cell
Metab., 19, 445-457.
53.Maio, N., and Rouault, T. A. (2015) Iron-sulfur
cluster biogenesis in mammalian cells: new insights into the molecular
mechanisms of cluster delivery, Biochim. Biophys. Acta,
1853, 1493-1512.
54.Bych, K., Kerscher, S., Netz, D. J., Pierik, A.
J., Zwicker, K., Huynen, M. A., Lill, R., Brandt, U., and Balk, J.
(2008) The iron-sulphur protein Ind1 is required for effective complex
I assembly, EMBO J., 27, 1736-1746.
55.Sheftel, A. D., Stehling, O., Pierik, A. J.,
Netz, D. J., Kerscher, S., Elsasser, H. P., Wittig, I., Balk, J.,
Brandt, U., and Lill, R. (2009) Human ind1, an iron-sulfur cluster
assembly factor for respiratory complex I, Mol. Cell. Biol.,
29, 6059-6073.
56.Lill, R., Hoffmann, B., Molik, S., Pierik, A. J.,
Rietzschel, N., Stehling, O., Uzarska, M. A., Webert, H., Wilbrecht,
C., and Muhlenhoff, U. (2012) The role of mitochondria in cellular
iron-sulfur protein biogenesis and iron metabolism, Biochim.
Biophys. Acta, 1823, 1491-1508.
57.Stehling, O., Wilbrecht, C., and Lill, R. (2014)
Mitochondrial iron-sulfur protein biogenesis and human disease,
Biochimie, 100, 61-77.
58.Guruharsha, K. G., Rual, J. F., Zhai, B.,
Mintseris, J., Vaidya, P., Vaidya, N., Beekman, C., Wong, C., Rhee, D.
Y., Cenaj, O., McKillip, E., Shah, S., Stapleton, M., Wan, K. H., Yu,
C., Parsa, B., Carlson, J. W., Chen, X., Kapadia, B., VijayRaghavan,
K., Gygi, S. P., Celniker, S. E., Obar, R. A., and Artavanis-Tsakonas,
S. (2011) A protein complex network of Drosophila melanogaster,
Cell, 147, 690-703.
59.Lim, S. C., Friemel, M., Marum, J. E., Tucker, E.
J., Bruno, D. L., Riley, L. G., Christodoulou, J., Kirk, E. P., Boneh,
A., DeGennaro, C. M., Springer, M., Mootha, V. K., Rouault, T. A.,
Leimkuhler, S., Thorburn, D. R., and Compton, A. G. (2013) Mutations in
LYRM4, encoding iron-sulfur cluster biogenesis factor ISD11, cause
deficiency of multiple respiratory chain complexes, Hum. Mol.
Genet., 22, 4460-4473.
60.Saha, P. P., Srivastava, S., Kumar, S. K. P.,
Sinha, D., and D’Silva, P. (2015) Mapping key residues of ISD11
critical for NFS1-ISD11 subcomplex stability: implications in the
development of mitochondrial disorder, COXPD19, J. Biol. Chem.,
290, 25876-25890.
61.Page, C. C., Moser, C. C., Chen, X., and Dutton,
P. L. (1999) Natural engineering principles of electron tunnelling in
biological oxidation-reduction, Nature, 402, 47-52.
62.Ohnishi, T. (1975) Thermodynamic and EPR
characterization of iron-sulfur centers in the NADH-ubiquinone segment
of the mitochondrial respiratory chain in pigeon heart, Biochim.
Biophys. Acta, 387, 475-490.
63.Nakamaru-Ogiso, E. (2012) Iron-sulfur clusters in
complex I, in A Structural Perspective on Respiratory Complex I
(Sazanov, L., ed.) Springer, The Netherlands, pp. 61-79.
64.Pohl, T., Bauer, T., Dorner, K., Stolpe, S.,
Sell, P., Zocher, G., and Friedrich, T. (2007) Iron-sulfur cluster N7
of the NADH:ubiquinone oxidoreductase (complex I) is essential for
stability but not involved in electron transfer, Biochemistry,
46, 6588-6596.
65.Tocilescu, M. A., Fendel, U., Zwicker, K., Drose,
S., Kerscher, S., and Brandt, U. (2010) The role of a conserved
tyrosine in the 49-kDa subunit of complex I for ubiquinone binding and
reduction, Biochim. Biophys. Acta, 1797, 625-632.
66.Tocilescu, M. A., Zickermann, V., Zwicker, K.,
and Brandt, U. (2010) Quinone binding and reduction by respiratory
complex I, Biochim. Biophys. Acta, 1797, 1883-1890.
67.Friedrich, T., Hellwig, P., and Einsle, O. (2012)
On the mechanism of the respiratory complex I, in A Structural
Perspective on Respiratory Complex I (Sazanov, L., ed.) Springer,
The Netherlands, pp. 23-59.
68.Hinchliffe, P., Carroll, J., and Sazanov, L. A.
(2006) Identification of a novel subunit of respiratory complex I from
Thermus thermophilus, Biochemistry, 45,
4413-4420.
69.Sazanov, L. A., and Hinchliffe, P. (2006)
Structure of the hydrophilic domain of respiratory complex I from
Thermus thermophilus, Science, 311, 1430-1436.
70.Ohnishi, T. (1998) Iron-sulfur
clusters/semiquinones in complex I, Biochim. Biophys. Acta,
1364, 186-206.
71.Hayashi, T., and Stuchebrukhov, A. A. (2010)
Electron tunneling in respiratory complex I, Proc. Natl. Acad. Sci.
USA, 107, 19157-19162.
72.Kmita, K., Wirth, C., Warnau, J.,
Guerrero-Castillo, S., Hunte, C., Hummer, G., Kaila, V. R., Zwicker,
K., Brandt, U., and Zickermann, V. (2015) Accessory NUMM (NDUFS6)
subunit harbors a Zn-binding site and is essential for biogenesis of
mitochondrial complex I, Proc. Natl. Acad. Sci. USA, 112,
5685-5690.
73.Ohnishi, T., and Salerno, J. C. (2005)
Conformation-driven and semiquinone-gated proton-pump mechanism in the
NADH-ubiquinone oxidoreductase (complex I), FEBS Lett.,
579, 4555-4561.
74.Zwicker, K., Galkin, A., Drose, S., Grgic, L.,
Kerscher, S., and Brandt, U. (2006) The Redox-Bohr group associated
with iron-sulfur cluster N2 of complex I, J. Biol. Chem.,
281, 23013-23017.
75.Yano, T., Dunham, W. R., and Ohnishi, T. (2005)
Characterization of the delta muH+-sensitive ubisemiquinone
species (SQ(Nf)) and the interaction with cluster N2: new insight into
the energy-coupled electron transfer in complex I, Biochemistry,
44, 1744-1754.
76.Nakamaru-Ogiso, E., Narayanan, M., and Sakyiama,
J. A. (2014) Roles of semiquinone species in proton pumping mechanism
by complex I, J. Bioenerg. Biomembr., 46, 269-277.
77.Sazanov, L. A. (2007) Respiratory complex I:
mechanistic and structural insights provided by the crystal structure
of the hydrophilic domain, Biochemistry, 46,
2275-2288.
78.Sun, F., Huo, X., Zhai, Y., Wang, A., Xu, J., Su,
D., Bartlam, M., and Rao, Z. (2005) Crystal structure of mitochondrial
respiratory membrane protein complex II, Cell, 121,
1043-1057.
79.Iverson, T. M. (2013) Catalytic mechanisms of
complex II enzymes: a structural perspective, Biochim. Biophys.
Acta, 1827, 648-657.
80.Van Vranken, J. G., Na, U., Winge, D. R., and
Rutter, J. (2015) Protein-mediated assembly of succinate dehydrogenase
and its cofactors, Crit. Rev. Biochem. Mol. Biol., 50,
168-180.
81.Iwata, S., Lee, J. W., Okada, K., Lee, J. K.,
Iwata, M., Rasmussen, B., Link, T. A., Ramaswamy, S., and Jap, B. K.
(1998) Complete structure of the 11-subunit bovine mitochondrial
cytochrome bc1 complex, Science, 281, 64-71.
82.Zhang, Z., Huang, L., Shulmeister, V. M., Chi, Y.
I., Kim, K. K., Hung, L. W., Crofts, A. R., Berry, E. A., and Kim, S.
H. (1998) Electron transfer by domain movement in cytochrome
bc1, Nature, 392, 677-684.
83.Akiba, T., Toyoshima, C., Matsunaga, T.,
Kawamoto, M., Kubota, T., Fukuyama, K., Namba, K., and Matsubara, H.
(1996) Three-dimensional structure of bovine cytochrome bc1
complex by electron cryomicroscopy and helical image reconstruction,
Nat. Struct. Biol., 3, 553-561.
84.Xia, D., Esser, L., Tang, W. K., Zhou, F., Zhou,
Y., Yu, L., and Yu, C. A. (2013) Structural analysis of cytochrome
bc1 complexes: implications to the mechanism of function,
Biochim. Biophys. Acta, 1827, 1278-1294.
85.Cooley, J. W. (2013) Protein conformational
changes involved in the cytochrome bc1 complex catalytic cycle,
Biochim. Biophys. Acta, 1827, 1340-1345.
86.Gurung, B., Yu, L., Xia, D., and Yu, C. A. (2005)
The iron-sulfur cluster of the Rieske iron-sulfur protein functions as
a proton-exiting gate in the cytochrome bc(1) complex, J.
Biol. Chem., 280, 24895-24902.
87.Iwata, S., Saynovits, M., Link, T. A., and
Michel, H. (1996) Structure of a water soluble fragment of the
“Rieske” iron-sulfur protein of the bovine heart
mitochondrial cytochrome bc1 complex determined by MAD phasing
at 1.5 Å resolution, Structure, 4, 567-579.
88.Link, T. A., and Iwata, S. (1996) Functional
implications of the structure of the “Rieske” iron-sulfur
protein of bovine heart mitochondrial cytochrome bc1 complex,
Biochim. Biophys. Acta, 1275, 54-60.
89.Smith, J. L., Zhang, H., Yan, J., Kurisu, G., and
Cramer, W. A. (2004) Cytochrome bc complexes: a common core of
structure and function surrounded by diversity in the outlying
provinces, Curr. Opin. Struct. Biol., 14, 432-439.
90.Esser, L., Gong, X., Yang, S., Yu, L., Yu, C. A.,
and Xia, D. (2006) Surface-modulated motion switch: capture and release
of iron-sulfur protein in the cytochrome bc1 complex, Proc.
Natl. Acad. Sci. USA, 103, 13045-13050.
91.Berry, E. A., De Bari, H., and Huang, L. S.
(2013) Unanswered questions about the structure of cytochrome
bc1 complexes, Biochim. Biophys. Acta, 1827,
1258-1277.
92.Borek, A., Kuleta, P., Ekiert, R., Pietras, R.,
Sarewicz, M., and Osyczka, A. (2015) Mitochondrial disease-related
mutation G167P in cytochrome b of rhodobacter capsulatus
cytochrome bc1 (S151P in human) affects the equilibrium
distribution of [2Fe-2S] cluster and generation of superoxide, J.
Biol. Chem., 290, 23781-23792.
93.Sanchez, E., Lobo, T., Fox, J. L., Zeviani, M.,
Winge, D. R., and Fernandez-Vizarra, E. (2013) LYRM7/MZM1L is a UQCRFS1
chaperone involved in the last steps of mitochondrial Complex III
assembly in human cells, Biochim. Biophys. Acta, 1827,
285-293.
94.Lehninger, A. L., Nelson, D. L., and Cox, M. M.
(2013) Lehninger Principles of Biochemistry, 6th Edn., W.H.
Freeman, New York.
95.Robbins, A. H., and Stout, C. D. (1989) The
structure of aconitase, Proteins, 5, 289-312.
96.Robbins, A. H., and Stout, C. D. (1989) Structure
of activated aconitase: formation of the [4Fe-4S] cluster in the
crystal, Proc. Natl. Acad. Sci. USA, 86, 3639-3643.
97.Lauble, H., Kennedy, M. C., Beinert, H., and
Stout, C. D. (1992) Crystal structures of aconitase with isocitrate and
nitroisocitrate bound, Biochemistry, 31, 2735-2748.
98.Talib, J., and Davies, M. J. (2016) Exposure of
aconitase to smoking-related oxidants results in iron loss and
increased iron response protein-1 activity: potential mechanisms for
iron accumulation in human arterial cells, J. Biol. Inorg.
Chem., 21, 305-317.
99.Myers, C. R., Antholine, W. E., and Myers, J. M.
(2010) The pro-oxidant chromium(VI) inhibits mitochondrial complex I,
complex II, and aconitase in the bronchial epithelium: EPR markers for
Fe-S proteins, Free Radic. Biol. Med., 49, 1903-1915.
100.Han, D., Canali, R., Garcia, J., Aguilera, R.,
Gallaher, T. K., and Cadenas, E. (2005) Sites and mechanisms of
aconitase inactivation by peroxynitrite: modulation by citrate and
glutathione, Biochemistry, 44, 11986-11996.
101.Beinert, H., and Kennedy, M. C. (1993)
Aconitase, a two-faced protein: enzyme and iron regulatory factor,
FASEB J., 7, 1442-1449.
102.Eisenstein, R. S. (2000) Iron regulatory
proteins and the molecular control of mammalian iron metabolism,
Annu. Rev. Nutr., 20, 627-662.
103.Hentze, M. W., and Kuhn, L. C. (1996) Molecular
control of vertebrate iron metabolism: mRNA-based regulatory circuits
operated by iron, nitric oxide, and oxidative stress, Proc. Natl.
Acad. Sci. USA, 93, 8175-8182.
104.Cairo, G., Recalcati, S., Pietrangelo, A., and
Minotti, G. (2002) The iron regulatory proteins: targets and modulators
of free radical reactions and oxidative damage, Free Radic. Biol.
Med., 32, 1237-1243.
105.Chen, X. J., Wang, X., Kaufman, B. A., and
Butow, R. A. (2005) Aconitase couples metabolic regulation to
mitochondrial DNA maintenance, Science, 307, 714-717.
106.Ferrer, M., Golyshina, O. V., Beloqui, A.,
Golyshin, P. N., and Timmis, K. N. (2007) The cellular machinery of
Ferroplasma acidiphilum is iron-protein-dominated, Nature,
445, 91-94.
107.White, M. F., and Dillingham, M. S. (2012)
Iron-sulphur clusters in nucleic acid processing enzymes, Curr.
Opin. Struct. Biol., 22, 94-100.
108.Boal, A. K., Yavin, E., and Barton, J. K.
(2007) DNA repair glycosylases with a [4Fe-4S] cluster: a redox
cofactor for DNA-mediated charge transport? J. Inorg. Biochem.,
101, 1913-1921.
109.Genereux, J. C., Boal, A. K., and Barton, J. K.
(2010) DNA-mediated charge transport in redox sensing and signaling,
J. Am. Chem. Soc., 132, 891-905.
110.Atta, M., Mulliez, E., Arragain, S., Forouhar,
F., Hunt, J. F., and Fontecave, M. (2010)
S-adenosylmethionine-dependent radical-based modification of biological
macromolecules, Curr. Opin. Struct. Biol., 20,
684-692.
111.Liu, H., Rudolf, J., Johnson, K. A., McMahon,
S. A., Oke, M., Carter, L., McRobbie, A. M., Brown, S. E., Naismith, J.
H., and White, M. F. (2008) Structure of the DNA repair helicase XPD,
Cell, 133, 801-812.
112.Wu, Y., Sommers, J. A., Suhasini, A. N.,
Leonard, T., Deakyne, J. S., Mazin, A. V., Shin-Ya, K., Kitao, H., and
Brosh, R. M., Jr. (2010) Fanconi anemia group J mutation abolishes its
DNA repair function by uncoupling DNA translocation from helicase
activity or disruption of protein–DNA complexes, Blood,
116, 3780-3791.
113.Landry, A. P., and Ding, H. (2014) The
N-terminal domain of human DNA helicase Rtel1 contains a redox active
iron-sulfur cluster, Biomed Res. Int., 285791.
114.Capo-Chichi, J. M., Bharti, S. K., Sommers, J.
A., Yammine, T., Chouery, E., Patry, L., Rouleau, G. A., Samuels, M.
E., Hamdan, F. F., Michaud, J. L., Brosh, R. M., Jr., Megarbane, A.,
and Kibar, Z. (2013) Identification and biochemical characterization of
a novel mutation in DDX11 causing Warsaw breakage syndrome, Hum.
Mutat., 34, 103-107.
115.Yeeles, J. T., Cammack, R., and Dillingham, M.
S. (2009) An iron-sulfur cluster is essential for the binding of broken
DNA by AddAB-type helicase-nucleases, J. Biol. Chem.,
284, 7746-7755.
116.Budd, M. E., Reis, C. C., Smith, S., Myung, K.,
and Campbell, J. L. (2006) Evidence suggesting that Pif1 helicase
functions in DNA replication with the Dna2 helicase/nuclease and DNA
polymerase delta, Mol. Cell. Biol., 26, 2490-2500.
117.Burgers, P. M., Stith, C. M., Yoder, B. L., and
Sparks, J. L. (2010) Yeast exonuclease 5 is essential for mitochondrial
genome maintenance, Mol. Cell. Biol., 30, 1457-1466.
118.Sparks, J. L., Kumar, R., Singh, M., Wold, M.
S., Pandita, T. K., and Burgers, P. M. (2012) Human exonuclease 5 is a
novel sliding exonuclease required for genome stability, J. Biol.
Chem., 287, 42773-42783.
119.Zhang, J., Kasciukovic, T., and White, M. F.
(2012) The CRISPR associated protein Cas4 is a 5′ to 3′ DNA
exonuclease with an iron-sulfur cluster, PLoS One, 7,
e47232.
120.Lemak, S., Beloglazova, N., Nocek, B., Skarina,
T., Flick, R., Brown, G., Popovic, A., Joachimiak, A., Savchenko, A.,
and Yakunin, A. F. (2013) Toroidal structure and DNA cleavage by the
CRISPR-associated [4Fe-4S] cluster containing Cas4 nuclease SSO0001
from Sulfolobus solfataricus, J. Am. Chem. Soc.,
135, 17476-17487.
121.Lemak, S., Nocek, B., Beloglazova, N., Skarina,
T., Flick, R., Brown, G., Joachimiak, A., Savchenko, A., and Yakunin,
A. F. (2014) The CRISPR-associated Cas4 protein Pcal_0546 from
Pyrobaculum calidifontis contains a [2Fe-2S] cluster:
crystal structure and nuclease activity, Nucleic Acids Res.,
42, 11144-11155.
122.Tuteja, N., and Tuteja, R. (2004) Prokaryotic
and eukaryotic DNA helicases. Essential molecular motor proteins for
cellular machinery, Eur. J. Biochem., 271, 1835-1848.
123.White, M. F. (2009) Structure, function and
evolution of the XPD family of iron-sulfur-containing
5′→3′ DNA helicases, Biochem. Soc. Trans.,
37, 547-551.
124.Wu, Y., and Brosh, R. M., Jr. (2012) DNA
helicase and helicase-nuclease enzymes with a conserved iron-sulfur
cluster, Nucleic Acids Res., 40, 4247-4260.
125.Suhasini, A. N., and Brosh, R. M., Jr. (2013)
DNA helicases associated with genetic instability, cancer, and aging,
Adv. Exp. Med. Biol., 767, 123-144.
126.Rudolf, J., Makrantoni, V., Ingledew, W. J.,
Stark, M. J., and White, M. F. (2006) The DNA repair helicases XPD and
FancJ have essential iron-sulfur domains, Mol. Cell, 23,
801-808.
127.Fan, L., Fuss, J. O., Cheng, Q. J., Arvai, A.
S., Hammel, M., Roberts, V. A., Cooper, P. K., and Tainer, J. A. (2008)
XPD helicase structures and activities: insights into the cancer and
aging phenotypes from XPD mutations, Cell, 133,
789-800.
128.Wolski, S. C., Kuper, J., Hanzelmann, P.,
Truglio, J. J., Croteau, D. L., Van Houten, B., and Kisker, C. (2008)
Crystal structure of the Fe-S cluster-containing nucleotide excision
repair helicase XPD, PLoS Biol., 6, e149.
129.Pugh, R. A., Honda, M., Leesley, H., Thomas,
A., Lin, Y., Nilges, M. J., Cann, I. K., and Spies, M. (2008) The
iron-containing domain is essential in Rad3 helicases for coupling of
ATP hydrolysis to DNA translocation and for targeting the helicase to
the single-stranded DNA–double-stranded DNA junction, J. Biol.
Chem., 283, 1732-1743.
130.Sommers, J. A., Banerjee, T., Hinds, T., Wan,
B., Wold, M. S., Lei, M., and Brosh, R. M., Jr. (2014) Novel function
of the Fanconi anemia group J or RECQ1 helicase to disrupt
protein–DNA complexes in a replication protein A-stimulated
manner, J. Biol. Chem., 289, 19928-19941.
131.Mishra, N. C. (1995) Molecular Biology of
Nucleases, CRC Press, Boca Raton.
132.Sisakova, E., Weiserova, M., Dekker, C.,
Seidel, R., and Szczelkun, M. D. (2008) The interrelationship of
helicase and nuclease domains during DNA translocation by the molecular
motor EcoR124I, J. Mol. Biol., 384, 1273-1286.
133.Wigley, D. B. (2013) Bacterial DNA repair:
recent insights into the mechanism of RecBCD, AddAB and AdnAB, Nat.
Rev. Microbiol., 11, 9-13.
134.Zheng, L., Zhou, M., Guo, Z., Lu, H., Qian, L.,
Dai, H., Qiu, J., Yakubovskaya, E., Bogenhagen, D. F., Demple, B., and
Shen, B. (2008) Human DNA2 is a mitochondrial nuclease/helicase for
efficient processing of DNA replication and repair intermediates,
Mol. Cell, 32, 325-336.
135.Duxin, J. P., Dao, B., Martinsson, P., Rajala,
N., Guittat, L., Campbell, J. L., Spelbrink, J. N., and Stewart, S. A.
(2009) Human Dna2 is a nuclear and mitochondrial DNA maintenance
protein, Mol. Cell. Biol., 29, 4274-4282.
136.Kang, Y. H., Lee, C. H., and Seo, Y. S. (2010)
Dna2 on the road to Okazaki fragment processing and genome stability in
eukaryotes, Crit. Rev. Biochem. Mol. Biol., 45,
71-96.
137.Taha, T. A., Kitatani, K., El-Alwani, M.,
Bielawski, J., Hannun, Y. A., and Obeid, L. M. (2006) Loss of
sphingosine kinase-1 activates the intrinsic pathway of programmed cell
death: modulation of sphingolipid levels and the induction of
apoptosis, FASEB J., 20, 482-484.
138.Abou-Ghali, M., and Stiban, J. (2015)
Regulation of ceramide channel formation and disassembly: insights on
the initiation of apoptosis, Saudi J. Biol. Sci., 22,
760-772.
139.Stiban, J., and Perera, M. (2015) Very long
chain ceramides interfere with C16-ceramide-induced channel formation:
a plausible mechanism for regulating the initiation of intrinsic
apoptosis, Biochim. Biophys. Acta, 1848, 561-567.
140.Wiley, S. E., Murphy, A. N., Ross, S. A., Van
der Geer, P., and Dixon, J. E. (2007) MitoNEET is an iron-containing
outer mitochondrial membrane protein that regulates oxidative capacity,
Proc. Natl. Acad. Sci. USA, 104, 5318-5323.
141.Kusminski, C. M., Holland, W. L., Sun, K.,
Park, J., Spurgin, S. B., Lin, Y., Askew, G. R., Simcox, J. A.,
McClain, D. A., Li, C., and Scherer, P. E. (2012) MitoNEET-driven
alterations in adipocyte mitochondrial activity reveal a crucial
adaptive process that preserves insulin sensitivity in obesity, Nat.
Med., 18, 1539-1549.
142.Landry, A. P., and Ding, H. (2014) Redox
control of human mitochondrial outer membrane protein MitoNEET [2Fe-2S]
clusters by biological thiols and hydrogen peroxide, J. Biol.
Chem., 289, 4307-4315.
143.Via, A., Ferre, F., Brannetti, B., Valencia,
A., and Helmer-Citterich, M. (2000) Three-dimensional view of the
surface motif associated with the P-loop structure: cis and
trans cases of convergent evolution, J. Mol. Biol.,
303, 455-465.
144.Lipper, C. H., Paddock, M. L., Onuchic, J. N.,
Mittler, R., Nechushtai, R., and Jennings, P. A. (2015) Cancer-related
NEET proteins transfer 2Fe-2S clusters to anamorsin, a protein required
for cytosolic iron-sulfur cluster biogenesis, PLoS One,
10, e0139699.
145.Golinelli-Cohen, M. P., Lescop, E., Mons, C.,
Goncalves, S., Clemancey, M., Santolini, J., Guittet, E., Blondin, G.,
Latour, J. M., and Bouton, C. (2016) Redox control of the human
iron-sulfur repair protein MitoNEET activity via its iron-sulfur
cluster, J. Biol. Chem., 291, 7583-7593.
146.Ferecatu, I., Goncalves, S., Golinelli-Cohen,
M. P., Clemancey, M., Martelli, A., Riquier, S., Guittet, E., Latour,
J. M., Puccio, H., Drapier, J. C., Lescop, E., and Bouton, C. (2014)
The diabetes drug target MitoNEET governs a novel trafficking pathway
to rebuild an Fe-S cluster into cytosolic aconitase/iron regulatory
protein 1, J. Biol. Chem., 289, 28070-28086.
147.Shulga, N., and Pastorino, J. G. (2014)
Mitoneet mediates TNFalpha-induced necroptosis promoted by exposure to
fructose and ethanol, J. Cell Sci., 127, 896-907.
148.Sohn, Y. S., Tamir, S., Song, L., Michaeli, D.,
Matouk, I., Conlan, A. R., Harir, Y., Holt, S. H., Shulaev, V.,
Paddock, M. L., Hochberg, A., Cabanchick, I. Z., Onuchic, J. N.,
Jennings, P. A., Nechushtai, R., and Mittler, R. (2013) NAF-1 and
mitoNEET are central to human breast cancer proliferation by
maintaining mitochondrial homeostasis and promoting tumor growth,
Proc. Natl. Acad. Sci. USA, 110, 14676-14681.
149.Shimizu, R., Lan, N. N., Tai, T. T., Adachi,
Y., Kawazoe, A., Mu, A., and Taketani, S. (2014) p53 directly regulates
the transcription of the human frataxin gene and its lack of regulation
in tumor cells decreases the utilization of mitochondrial iron,
Gene, 551, 79-85.
150.Shakoury-Elizeh, M., Protchenko, O., Berger,
A., Cox, J., Gable, K., Dunn, T. M., Prinz, W. A., Bard, M., and
Philpott, C. C. (2010) Metabolic response to iron deficiency in
Saccharomyces cerevisiae, J. Biol. Chem., 285,
14823-14833.
151.Yang, Z., Wang, W. E., and Zhang, Q. (2013)
CIAPIN1 siRNA inhibits proliferation, migration and promotes apoptosis
of VSMCs by regulating Bcl-2 and Bax, Curr. Neurovasc. Res.,
10, 4-10.
152.Banci, L., Ciofi-Baffoni, S., Mikolajczyk, M.,
Winkelmann, J., Bill, E., and Pandelia, M. E. (2013) Human anamorsin
binds [2Fe-2S] clusters with unique electronic properties, J. Biol.
Inorg. Chem., 18, 883-893.
153.Netz, D. J., Stumpfig, M., Dore, C.,
Muhlenhoff, U., Pierik, A. J., and Lill, R. (2010) Tah18 transfers
electrons to Dre2 in cytosolic iron-sulfur protein biogenesis, Nat.
Chem. Biol., 6, 758-765.
154.Vernis, L., Facca, C., Delagoutte, E., Soler,
N., Chanet, R., Guiard, B., Faye, G., and Baldacci, G. (2009) A newly
identified essential complex, Dre2-Tah18, controls mitochondria
integrity and cell death after oxidative stress in yeast, PLoS
One, 4, e4376.