Coupling of Translation Initiation and Termination Does Not Depend on the Mode of Initiation
E. A. Sogorin, G. K. Selikhanov, and S. Ch. Agalarov*
Institute of Protein Research, Russian Academy of Sciences, 142290 Pushchino, Moscow Region, Russia; E-mail: sultan@vega.protres.ru* To whom correspondence should be addressed.
Received February 21, 2017; Revision received March 27, 2017
Recently we described a novel phenomenon observed during eukaryotic translation in a cell-free system: the coupling of initiation and termination on different mRNA molecules. Here we show that the phenomenon does not depend on a special mode of initiation. The mRNAs with certain leader sequences known to require different determinants for successful initiation were examined. Even in a case of using the intergenic internal ribosome entry site (IRES) of cricket paralysis virus RNA as the leader sequence, while no initiation factors are required, the effect of coupling is well expressed, including trials in the presence of hippuristanol as an inhibitor of eIF4A. Thus, the effect persists in the absence of scanning and does not depend on initiator tRNA and eIF2. The results suggest that the initiation factors are not involved in the coupling mechanism.
KEY WORDS: translation initiation, translation termination, initiation factors, hippuristanolDOI: 10.1134/S0006297917070069
Abbreviations: IRES, internal ribosome entry site; UTR, untranslated region.
In the process of protein synthesis, the translating ribosomes go
through a translation cycle consisting of initiation, elongation, and
termination phases, or stages. Every full cycle usually results in the
reading out of the coding sequence of the mRNA and ends by the
synthesis of a full-length polypeptide. The termination process
involves stop-codon recognition and the hydrolysis of the
peptidyl-tRNA, which leads to the release of the full-length
polypeptide from the ribosome [1, 2]. Earlier, using continuous recording of
luciferase-generated luminescence in situ [3], it was shown that the addition of exogenous mRNA
into the working translation system induced the immediate release of
the complete (terminated) protein from the ribosomes [4]. The effect did not depend on the coding
specificity of the added mRNA. However, it depended on the
translational activity of the added mRNA: direct correlation between
translational activity of mRNAs and output of the protein release was
revealed. The translational activity, in turn, primarily depended on
the efficiency of translation initiation. It is also important to note
that exactly the translational activity of the mRNA, no matter what it
is due to (the presence of a cap structure or sufficient concentration
of uncapped mRNAs), was responsible for the occurrence of the effect
and its extent. On the other hand, the phenomenon was also strictly
dependent on the presence of the stop codon in the initial translated
mRNA (no termination – no effect). These observations implied the
coupling of the two processes: initiation on the newly added mRNAs and
termination on the “old” mRNAs, using which the translation
system was run. At that, the coupling obeys the rule: the more
effective initiation, the more expressed termination.
Considering the molecular mechanism of the coupling phenomenon, the question has been raised: are the initiation factors involved in the process? In our recent study, when 5′-UTR of the mRNA encoding for protein obelin (light-emitting protein of the hydroid polyp Obelia longissima) was used in experiments, it was found that it did not need the cap structure and the factors eIF4A and eIF4B to initiate translation [5]. It has been shown that the internal ribosome entry site (IRES) of the cricket paralysis virus was capable of initiating translation without any initiation factors [6]. Using these and some other leader sequences, it was demonstrated that the immediate release of the complete (terminated) protein from ribosomes is not dependent on the mode of translation initiation. It can be concluded that initiation factors probably do not participate in the mechanism of the coupling of initiation and termination.
MATERIALS AND METHODS
Plasmids. Plasmid pTZ10ΩLuc containing the firefly luciferase-coding region with the omega sequence was described in [7]. To obtain the RNA containing the leader sequence of the mRNA encoding obelin, the plasmid construct pObeLucTMV was used [8]. The construct containing the firefly luciferase-coding region with the β-globin mRNA leader has been described [9]. The plasmid encoding the intergenic IRES of the cricket paralysis virus [10] was kindly provided by S. E. Dmitriev.
mRNA preparations. All procedures for the transcription and purification of mRNAs were described in detail elsewhere [11]. The reaction was performed at 37°C for 2 h. The purity of the polyribonucleotides obtained was verified by electrophoresis in 6% polyacrylamide gel with 7 M urea. Capping of the mRNAs was performed using the Vaccinia Capping System (NEB, USA), with following purification in MicroSpinTM G-50 columns (GE Healthcare, GB).
In situ monitoring of luciferase synthesis. The method was described in detail previously [7]. Translation was performed in a cell-free translation system based on wheat germ extract using the batch format according to a published protocol [12]. The volume of the reaction mixture was 10 µl. The translation system was started by addition of the mRNA to be translated; the reaction mixture was incubated at 25°C in the temperature-controlled cell of the luminometer (Hemilum-12; Institute of Cell Biophysics, Russian Academy of Sciences). The intensity of light emission was measured continuously (every 2.5 s) by collecting the streaming data with the computer as a kinetic curve. In the case of the post-start addition of mRNA, the luminometer was temporarily turned off, the mRNA was added into the cell, and the mixture was stirred. Then, as soon as the samples were returned into the luminometer, it was switched on again and the collection of luminescence data was continued.
RESULTS AND DISCUSSION
To retrace the process of translation, we used the method of the cell-free synthesis of firefly luciferase, which allows measuring enzymatic activity directly in the reaction mixture [3]. The enzyme catalyzes the ATP-dependent conversion of the luciferin substrate to its oxyform accompanied by emission of light. Hence, the activity of the newly synthesized luciferase can be continuously recorded during incubation of the translation mixture in a luminometer cell.
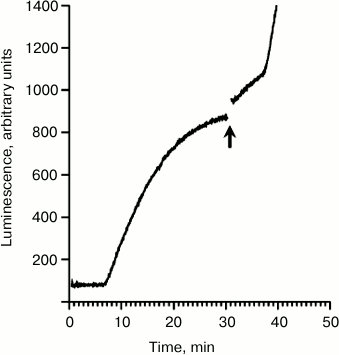
Figure 1 demonstrates the time course curve obtained by continuous monitoring of the luciferase activity in the wheat germ system. The translation was started by the addition of luciferase-encoding mRNA with the omega leader sequence (5′-UTROmega-Luc). This mRNA served as a basic template in our previous experiments when the effect of the coupling of initiation and termination was first described [4]. The lag time between the start of translation and the appearance of the first active product is ~8 min, while the plateau is reached soon after 30 min (Fig. 1). However, if extra Luc-mRNA (5′UTRObe-Luc) with leader sequence of mRNA encoding obelin was added to the translation mixture shortly before reaching the plateau, then two jumps of the luminescence intensity were observed. The observed rise of luminescence intensity implies the release of the active enzyme, since luciferase displays its activity only upon leaving the ribosome [3]. The first instant rise of luminescence intensity as a response to the addition of vacant mRNAs occurs for substantially less time than it is required for the synthesis of full-sized protein chains, or transit time (8 min in this case, it can vary up to 11 min). This implies that the luciferase released during the first rise was already pre-synthesized, i.e. its polypeptide chain was already complete or almost complete. The interval between the addition of extra mRNA and the second rise corresponds well to the transit time for translating ribosomes, implying that the second rise reflects the beginning of new translation cycles. Thus, the supplied free mRNA causes two events in the working translation system. The first is the instant release of completed polypeptides, i.e. the successful completion of the protein synthesis (translation termination) on mRNAs, using which the translation system was started. Second, the resumption of the synthesis of new polypeptide chains detected after the transit time, implying de novo initiation on the newly added mRNAs. Correspondingly, the effect has been called coupling of translation initiation and termination [4].
Fig. 1. Time course of the synthesis of functionally active luciferase in the cell-free translation system. The translation was initially started by the addition of the mRNA 5′-UTROmega-Luc (see text) up to its final concentration of 100 pmol/ml. The mRNA 5′-UTRObe-Luc was added into the active translation system just before the plateau stage as marked by the arrow up to the final concentration of 500 pmol/ml.
Earlier, it was demonstrated that the phenomenon of the coupling was independent of whether capped or uncapped (but well translatable!) mRNAs were used [4]. In that investigation, mRNAs with the capped 5′-UTR of the globin mRNA and the uncapped omega leader sequence as the newly added templates were studied. The effect of the coupling was well expressed in both cases. Since the initiation on these leader sequences, as known, can occur at different sets of initiation factors [13, 14], it could be assumed that the coupling of translation initiation and termination in some way does not depend on the initiation mode. The experiment when the 5′-UTR of the mRNA encoding for protein obelin was used as a leader sequence causing termination (Fig. 1) supports the idea. As recently shown, the leader sequence is capable of initiating at another minimal set of initiation factors than the omega leader and 5′-UTR of the globin mRNA [5].
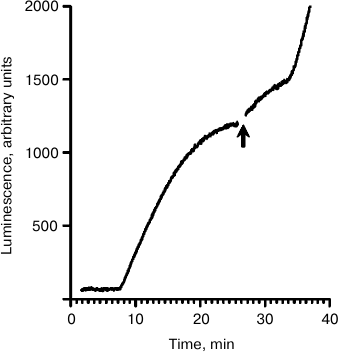
The mechanism of translation initiation on mRNAs containing the intergenic IRES of the cricket paralysis virus stands apart from all known initiation modes. The IRES bypasses the dependence of translation on all initiation factors and even Met-tRNAiMet [6]. In the used mRNA containing the IRES, there was no AUG codon [10]; therefore, no other way for translation initiation was possible – the eIF2-independent one. In other words, this mRNA is not able to use the standard pathway even in a complete cell lysate. Figure 2 demonstrates the effect of the addition of the mRNA containing the IRES into the working translation system. The translation was started by the addition of the luciferase-encoding mRNA with the omega leader sequence (5′-UTROmega-Luc). Just before the plateau, Luc-mRNA with the IRES sequence (5′-UTRCrPV-Luc) was added into the translation system. After the addition, two sequential rises of the luminescence intensity with interval corresponding to the transit time (~8 min) can be seen again. This implies that the coupling of initiation at 5′-UTRCrPV-Luc and termination at 5′-UTROmega-Luc occurred. As the 5′-UTRCrPV-Luc construct initiates translation without any initiation factors, it seems to be most probable that they are not also involved in the coupling of initiation and termination.
Fig. 2. Time course of the synthesis of functionally active luciferase in the cell-free translation system. The translation was initially started by the addition of mRNA 5′-UTROmega-Luc (see text) up to its final concentration of 100 pmol/ml. The mRNA 5′-UTRCrPV-Luc was added into the active translation system just before the plateau stage as marked by the arrow, up to the final concentration of 500 pmol/ml.
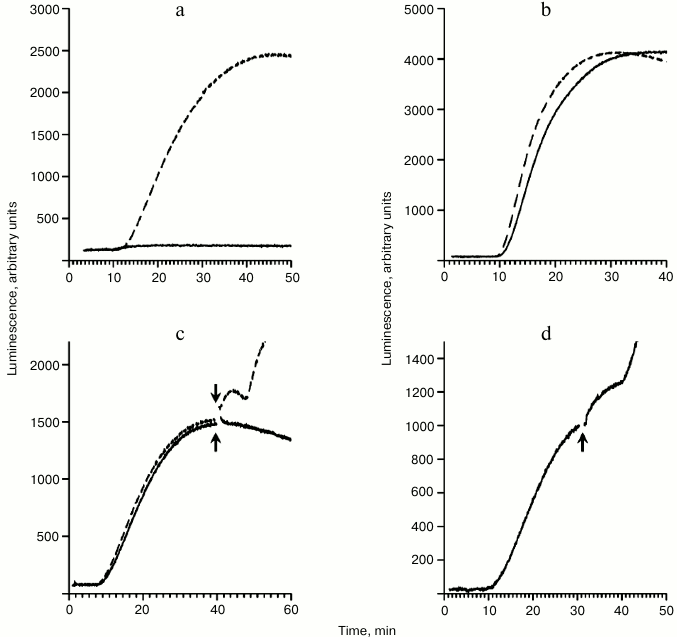
The next series of experiments was performed in the presence of the polyhydroxysteroid hippuristanol. Hippuristanol was found to inhibit translation initiation by blocking the activity of the initiation factor eIF4A [15]. Thus, in the presence of hippuristanol initiation factor eIF4A is definitely not involved in the process. Figure 3a shows the effect of hippuristanol on translation of mRNA with capped 5′-UTR of the globin mRNA (cap-5′-UTRβGlobin-Luc). It is clear that hippuristanol completely inhibits translation with cap/factor-dependent initiation. Unlike cap-5′-UTRβGlobin-Luc, hippuristanol does not influence translation of the 5′-UTRCrPV-Luc (Fig. 3b), expectedly implying irrelevance of eIF4A in the initiation process of this template. Figure 3c clearly displays how the absence of translation activity of added mRNA (due to the presence of hippuristanol) fully deprives it of the capability to induce the luciferase release. In addition, the effect of the coupling of initiation and termination in the presence of hippuristanol is shown in Fig. 3d. In this case, the translation was started by the addition of 5′-UTRCrPV-Luc and the same mRNA was added before the plateau stage. The result clearly implicates that eIF4A is not involved in the process of the coupling.
Fig. 3. Time courses of the synthesis of functionally active luciferase in the absence and presence of hippuristanol. When appropriate, the hippuristanol was added to its final concentration of 0.01 mM 2 min before the system started working. a) Translation of the cap-5′-UTRβGlobin-Luc mRNA (50 pmol/ml) in the presence of hippuristanol (solid curve) and without hippuristanol (dashed curve). b) Translation of the 5′-UTRCrPV-Luc mRNA (500 pmol/ml) in the presence of hippuristanol (solid curve) and without hippuristanol (dashed curve). c) The translation was initially started by the addition of the 5′-UTRCrPV-Luc mRNA (200 pmol/ml) in the presence of hippuristanol (solid curve) and without hippuristanol (dashed curve). The cap-5′-UTRβGlobin-Luc mRNA (50 pmol/ml) was added into the translation system just before the plateau stage as marked by the arrows. d) The translation was started by the addition of the 5′-UTRCrPV-Luc mRNA (200 pmol/ml) in the presence of hippuristanol. The same mRNA (500 pmol/ml) was added into the translation system just before the plateau stage as marked by the arrow.
Thus, exploring the phenomenon of the coupling of translation initiation and termination, we have examined mRNAs with several leaders known to have different mechanisms of initiation: cap/factor-dependent (cap-5′-UTRβGlobin), cap/eIF4F/ATP-independent (5′-UTROmega), cap-independent, eIF4F/ATP-dependent (5′-UTRObe), and cap/factor-independent, internal (5′-UTRCrPV). The effect of the coupling persisted in all investigated cases. We conclude that the coupling does not depend on the initiation mode. In addition, the experiments with mRNA containing the IRES of the cricket paralysis virus including trials in the presence of hippuristanol suggest that initiation factors are not involved in the mechanism of the coupling. Furthermore, translation initiation on mRNA with the IRES bypasses the scanning stage and includes only the last steps of conventional initiation – binding of the small subunit with start codon and recruitment of the large subunit to the initiation complex. We suggest that the coupling of initiation and termination is somehow related to structural reorganization of the translation machinery during formation of the full 80S ribosome under initiation. At the same time, the idea of a possible role of protein ABCE1 in the coupling mechanism first described in the previous work remains acceptable [4]. The idea is that being in bound form within the pre-initiation 43S complex, protein ABCE1 is not able to promote the processes of termination or recycling of translating ribosomes, this leading to their stalling. The addition of free exogenous mRNA induces the involvement of the pre-initiation 43S complexes in the translation initiation process and thus leads to the release of ABCE1. Afterwards, ABCE1 promotes termination or recycling of the stalled ribosomes.
Albeit the molecular mechanism of the coupling of translation initiation and termination is still unclear, the data presented here are unambiguously helpful for understanding the remarkable phenomenon.
Acknowledgments
The authors are grateful to A. S. Spirin for his support, constant interest, and valuable advice, to V. A. Kolb for reading the manuscript and critical comments, and to S. E. Dmitriev (Lomonosov Moscow State University) for providing us with the construct encoding the IRES of the cricket paralysis virus.
This work was supported by the Russian Foundation for Basic Research (project No. 15-04-015-25a) and by a grant of the program “Molecular and Cell Biology” of the Presidium of the Russian Academy of Sciences.
REFERENCES
1.Jackson, R. J., Hellen, C., and Pestova, T. V.
(2012) Termination and post-termination events in eukaryotic
translation, Adv. Protein Chem. Struct. Biol., 86,
45-93.
2.Dever, T. E., and Green, R. (2012) The elongation,
termination, and recycling phases of translation in eukaryotes, Cold
Spring Harb. Perspect. Biol., 4, 1-16.
3.Kolb, V. A., Makeyev, E. V., and Spirin, A. S.
(1994) Folding of firefly luciferase during translation in a cell-free
system, EMBO J., 13, 3631-3637.
4.Sogorin, E. A., Agalarov, S. C., and Spirin, A. S.
(2016) Inter-polysomal coupling of termination and initiation during
translation in eukaryotic cell-free system, Sci. Rep., 6,
24518.
5.Sakharov, P. A., and Agalarov, S. C. (2016) Free
initiation factors eIF4A and eIF4B are dispensable for translation
initiation on uncapped mRNAs, Biochemistry (Moscow), 81,
1198-1204.
6.Wilson, J. E., Pestova, T. V., Hellen, C. U., and
Sarnow, P. (2000) Initiation of protein synthesis from the A site of
the ribosome, Cell, 102, 511-520.
7.Alekhina, O. M., Vassilenko, K. S., and Spirin, A.
S. (2007) Translation of non-capped mRNAs in a eukaryotic cell-free
system: acceleration of initiation rate in the course of polysome
formation, Nucleic Acids Res., 35, 6547-6559.
8.Kopeina, G. S., Afonina, Z. A., Gromova, K. V.,
Shirokov, V. A., Vasiliev, V. D., and Spirin, A. S. (2008) Step-wise
formation of eukaryotic double-row polyribosomes and circular
translation of polysomal mRNA, Nucleic Acids Res., 36,
2476-2488.
9.Lyabin, D. N., Eliseeva, I. A., and Ovchinnikov, L.
P. (2012) YB-1 synthesis is regulated by mTOR signaling pathway,
PLoS One, 7, 1-11.
10.Prokhorova, I. V., Akulich, K. A., Makeeva, D.
S., Osterman, I. A., Yusupov, M. M., and Dmitriev, S. E. (2016)
Amicoumacin A induces cancer cell death by targeting the eukaryotic
ribosome, Sci. Rep., 6, 27720.
11.Sogorin, E. A., Agalarov, S. C., and Spirin, A.
S. (2015) Formation of new polysomes on free mRNAs in a cell-free
translation systems is accompanied by partial disassembly of previously
formed polysomes, Biochemistry (Moscow), 80,
1327-1330.
12.Shirokov, V. A., Kommer, A., Kolb, V. A., and
Spirin, A. S. (2007) Continuous-exchange protein-synthesizing systems,
Methods Mol. Biol., 375, 19-55.
13.Agalarov, S. C., Sakharov, P. A., Fattakhova, D.
K., Sogorin, E. A., and Spirin, A. S. (2014) Internal translation
initiation and eIF4F/ATP-independent scanning of mRNA by eukaryotic
ribosomal particles, Sci. Rep., 4, 4438.
14.Pestova, T. V., and Kolupaeva, V. G. (2002) The
roles of individual eukaryotic translation initiation factors in
ribosomal scanning and initiation codon selection, Genes Dev.,
16, 2906-2922.
15.Bordeleau, M.-E., Mori, A., Oberer, M.,
Lindqvist, L., Chard, L. S., Higa, T., Belsham, G. J., Wagner, G.,
Tanaka, J., and Pelletier, J. (2006) Functional characterization of
IRESes by an inhibitor of the RNA helicase eIF4A, Nat. Chem.
Biol., 2, 213-220.