REVIEW: Antibiotics from Marine Bacteria
V. A. Stonik1,a*, T. N. Makarieva1, and L. K. Shubina1
1Elyakov Pacific Institute of Bioorganic Chemistry, Far Eastern Branch of the Russian Academy of Sciences (PIBOC), 690022 Vladivostok, Russia* To whom correspondence should be addressed.
Received July 3, 2020; Revised August 5, 2020; Accepted August 5, 2020
This review discusses main directions and results of the studies on antibiotics produced by bacteria living in the marine environment. In recent years many obligate marine species and strains were studied, diverse metabolites were isolated, and their chemical structures were elucidated. Among them here were natural compounds toxic against tumor cells, pathogenic bacteria, viruses, and malaria plasmodial species; these compounds often had no analogues among the natural products of terrestrial origin. Some isolated compounds form a basis of active ingredients in medicinal preparations used in clinic practice, while others are under different stages of preclinical or clinical studies. Much attention has been paid in recent years to producers of marine-derived antibiotics isolated from the deep-sea habitats, from the surface of marine invertebrates and algae, as well as from symbiotic microorganisms.
KEY WORDS: marine bacteria, antibiotics, cytotoxic effect, antibacterial effect, antifungal effect, antiviral effect, antiparasitic effectDOI: 10.1134/S0006297920110073
INTRODUCTION
The discovery and widespread use of antibiotics had been of paramount importance for the fate of mankind and significantly reduced the number of deaths of epidemics that claimed lives of millions of people for many centuries. It is believed that this was one of the main causes for the increase in human life expectancy. For example, in Russia at the end of the 19th century, it was about 30 years, by 1961-1962 it had grown to 67.85 years, and now life expectancy is about 73.4 years (2018). However, the main natural source of antibiotics, soil microorganisms, has now significantly exhausted its potential, and the number of new antibiotics isolated from soil bacteria is decreasing.
At the same time, the search for new antibiotics remains an important task due to the increasing number of cases of severe infections caused by various pathogens, primarily drug-resistant strains. As a result, the number of patients with drug-resistant diseases is increasing. In 2017, 10 million people were diagnosed with tuberculosis, and 1.6 million people (including 0.3 million people with HIV) died from this disease [1]. In 2008 a group of pathogens associated with the severe drug-resistant infections was identified, which was named “ESKAPE BUGS” (acronym of the Latin names of the following antibiotic insensitive pathogens: Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumanii, Pseudomonas aeruginosa, and Enterobacter spp.) referring to their ability to ESCAPE the effects of the commonly used antibiotics. Later, properties of these pathogens were described in more detail [2]. The threat of infection with antibiotic-resistant strains of several other pathogenic bacteria, including Helicobacter pylori, Campylobacter spp., Neisseria gonorrhoeae, and others also increased in recent years [3].
The urgent need to find new antibiotics forced scientists to pay attention to the natural sources of these biologically active substances that are difficult to study, especially such as marine microorganisms – bacteria and microscopic fungi – that live under extreme conditions, namely to the so-called extremophiles (barophiles, psychrophiles, thermophiles, etc.), as well as epiphytic and symbiotic microbial flora of invertebrates and algae. Using new opportunities to search for antibiotics, including descent underwater vehicles and specialized ships, scientists have found antibacterial, antifungal substances and compounds that inhibit pathogenic protozoa in such non-traditional producers of antibiotics as animals, especially marine invertebrates, inhabitants of underwater hot springs, searched for these substances in the Arctic and Antarctica, at the maximum depths of the ocean, in deep caves and deserts. As a result, a series of substances with anti-pathogenic properties was identified, which can be considered as leader compounds for design of new drugs.
To date, several review articles have been published on various aspects of the search for antibiotics isolated from marine microorganisms (see [4-10] for example) describing more than a hundred of such compounds. The reviews concerning isolated metabolites that inhibit methicillin-resistant S. aureus [11] and vancomycin-resistant enterococci were also published [11, 12].
Although a significant number of marine microorganisms have not yet been cultured, other marine microorganisms have been studied and appeared to be quite different in their physiological and morphological features when compared with their terrestrial counterparts. These peculiarities help them to adapt and survive in the seas and oceans. Among marine microorganisms, there are many previously unknown species, genera, and even families of bacteria, and their discovery has greatly changed modern microbiology. For instance, several hundred new species of bacteria were validly described by scientists from our Institute under the leadership of Professor V. V. Mikhailov – corresponding member of the Russian Academy of Sciences. Like other marine bacteria, they produce a wide variety of bioactive metabolites [10, 13]. The first accurately described new species of heterotrophic bacteria in the Russian Far East region was Alteromonas fuliginea (type strain KMM 216T), inhabiting waters of the Peter the Great Bay (1995). Later, this species was assigned to the newly established genus Pseudoalteromonas and included in the species Pseudoalteromonas citrea. Over the next ten years, Marinomonas primoryensis sp. nov. (named in honor of the Primorye Region, Russia), Vitellibacter vladivostokensis gen. nov., sp. nov. (named in honor of Vladivostok), Arenibacter troitsensis sp. nov. (named in honor of Trinity Bay, in which waters this species lives, and on the shores of which the Marine experimental station (MES) of the PIBOC is located), Mesonia algae gen. nov., sp. nov. (named in honor of MES), Salinibacterium amurskyense gen. nov., sp. nov. (these bacteria were isolated from a sample of seawater from the Amursky Bay), Reinekea marinisedimentorum gen. nov., sp. nov. (named in honor of the Reinecke Island near Vladivostok, in waters of which the corresponding microorganism was found in the sandy soil at a shallow depth) and others were described. Only during the period from 1995 to 2005, more than 100 new species, about 40 new genera, and one new family of marine bacteria were discovered by the scientists working at the Microbiology laboratory of PIBOC [13]. Some of them produce a variety of bioactive compounds, including antibiotics [14]. To date, the number of new species and genera in the collection of marine microorganisms (acronym is KMM) of the PIBOC has increased significantly.
However, not so long ago, in 1996, only 7 genera of bacteria were considered exclusively marine, such as Oceanospirillum, Marinomonas, Planococcus, Listonella, Leucothrix, Platobacterium, and Prochloron [4]. Many of them are associated with marine fish, invertebrates, or algae. Later, the situation changed and the number of such taxa increased. Moreover, taxa of a higher taxonomic rank were found, all the representatives of which were obligate marine organisms. On the other hand, even in the cases when bacteria isolated from the sea water or sediments were found to belong to species previously isolated from the terrestrial biological sources, they could not be considered simply as washed away from the land and preserved all their properties in the new environment. Adaptation to the marine conditions often led to the selection of such metabolic pathways that generated previously unknown metabolites. This explains the significant biochemical diversity in these microorganisms and contributes to their study as new sources of antibiotics.
Actinobacteria are the most known bacterial producers of antibiotics. They belong to the family Actinomycetaceae, which includes the genera Streptomyces, Actinobaculum, Acanabacterium, and others. Most of these Gram-positive bacteria produce compounds with unique chemical structures that are toxic against other bacteria, including pathogenic ones. More than 60% of all antibiotics produced by the pharmaceutical industry were obtained by culturing actinobacteria [15, 16]. These include streptomycin, actinomycin, polyenes, tetracyclines, macrolides, and other medications. It is believed that each strain of actinobacteria, due to its genetic potential, is capable of synthesizing 15-25 different secondary metabolites. It was estimated that about 10 thousand new bioactive substances of various chemical natures could be found in actinobacteria [16].
For many decades, terrestrial sources, mainly soil microorganisms, have been used as a source for searching and designing new antibiotics. Although as early as in 1969, there was a report about the first mycelium-forming actinobacteria from marine bottom sediments [17]. Moreover, it has been suggested that marine antibiotics, including those isolated from symbiotic microorganisms of marine invertebrates, will have an advantage in creating new drugs [17, 18].
Among the various genera of actinobacteria, the genus Streptomyces is of particular interest, because it is known as a source of more than half of all antibiotics currently used in clinical practice. 5200 genomes of these actinomycetes are registered in the Genome Online Database [19]. Interestingly, new species of this genus have been discovered in recent years, which can be considered exclusively marine streptomycetes [20]. Moreover, 5 new genera of marine actinomycetes have been recently described: Marinactinospora [21], Jonesia [22], Salinibacterium [23], Sciscionella [24], and Salinispora [25]. Many new bacteria, belonging to other taxonomic groups were also discovered, and highly active secondary metabolites were found in them.
In this short review, we will provide information about the main directions of the search for marine-derived antibiotics and illustrate these directions only with a small number of examples of promising marine-derived antibiotics, grouping them by their inhibitory effects against tumor cells, pathogenic bacteria, fungi, and other pathogens.
CYTOTOXIC ANTIBIOTICS FROM BACTERIA ISOLATED FROM MARINE
BIOLOGICAL SOURCES
The search for marine antibiotics that exhibit an inhibitory effect against tumor cells has been actively carried out in several countries in recent years, especially in the United States and Japan. Salinispora bacteria have attracted much attention from microbiologists, bioorganic chemists, and pharmacologists. Their cultivation was first performed in 1989 after isolation from the samples of bottom sediments collected at a depth of 1100 meters. These bacteria, which originally were assigned to the genus Micromonospora, did not grow if sea water was replaced with ordinary deionized water in the medium. As Salinispora, they were described in 2005 [25]. After the cultivation technique was improved, Salinispora spp. were also found in some marine invertebrates (sponges, ascidia) and in algae. Bacteria of this genus turned out to be obligately marine species, and their further study led to the discovery of several antibiotics that exhibit strong antitumor effect [26].
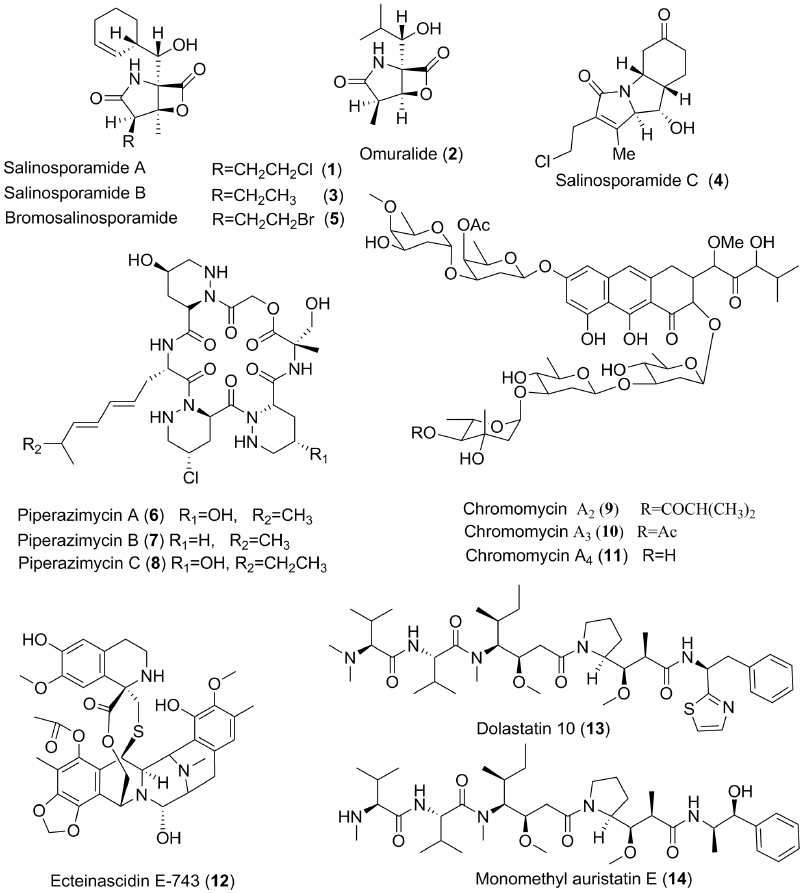
The first group of interesting metabolites from this species were obtained from the bacteria isolated from a pre-heated sample of bottom sediments. The main obtained substance was called salinosporamide A (1) [27]. This new antibiotic has a skeletal system consisting of γ-lactam and β-lactone (Fig. 1) and demonstrates cytotoxic properties against tumor cells. Although its cyclic system is identical to that of omuralide (2), isolated by Japanese microbiologists in 1991 from Streptomyces lactacystennaosus, the compound differs significantly from the omuralide (2) in its substituents and their positions. Salinosporamide A proved to be a powerful and selective inhibitor of proteasomes; it is approximately 35-fold more active in this test than omuralide. The antibiotic inhibits cell lines of lung cancer NCI-H226, brain tumors SF-539, melanoma SK-MEL-28 and MDA-MB-43, with IC50 values (concentrations that inhibit cells by 50%) in these tests being less than 10 nM. Salinosporamide proved to be an efficient anti-cancer agent in preclinical trials, and is undergoing human clinical trials as a preparation against melanoma. The corresponding drug called Marizomib is under development by “Nereus Pharmaceutical” (San Diego, USA) under a license obtained from the University of California, San Diego.
Fig. 1. Some cytotoxic metabolites of marine bacteria.
Different strains of Salinispora spp. produce also other antibiotics. For instance, the study of the S. tropica, strain CNB-392, led to isolation of salinosporamides B (3) and C (4), while replacement of NaCl with NaBr in the medium produced the compound (5) (Fig. 1), which has a bromine atom instead of chlorine in the side chain and retains cytotoxic properties [28, 29].
Cyclic peptides from the bacterium Streptomyces sp. (strain CNQ-593) from marine bottom sediments collected at a depth of 20 m near the Guam Island are also considered promising for cancer treatment. These substances were called piperazimycins and have formulae (6-8) (Fig. 1) [30]. Piperazimycins show strong toxic properties against tumor cells. The average cell growth-inhibitory concentration (GC50) after testing on 60 tumor cell lines, provided by the National Cancer Institute, USA, was approximately 100 nM.
In many cases, highly active metabolites from marine bacteria exhibit both cytotoxic and antibacterial properties, although sensitivity of the tumor cells and pathogenic bacteria to them could be different. For example, new derivatives of aureolic acid, chromomycins A2-A4 (9-11) (Fig. 1), obtained from Streptomyces sp. (strain KKM 9048, which was isolated from a sediment sample collected in the North-western part of the Sea of Japan), showed strong antibacterial action against Enterococcus faecium, Staphylococcus aureus, S. epidermis, and Bacillus subtilis. However, their inhibitory concentrations against tumor cells were even lower, especially those that inhibit formation of microcolonies of RPMI-7951 and SK-Mel-28 tumor cells (10-20 nM) [31].
The marine-derived isoquinoline derivative, ecteinascidin-743 (ET-743) (12) (Fig. 1), became the active substance of the new anticancer drug Trabectedin (Yondelis R), developed by the Spanish company “PharmaMar” (manufacturer “Baxter Oncology GmbH”, Germany). This substance itself was discovered in the USA at the University of Illinois by Prof. K. Rinehart. He was able to isolate it from the ascidian Ecteinascidia turbinata and demonstrate its very high antitumor activity [32]. The Nobel Prize laureate E. J. Corey and collaborators synthesized compound 12, but later a semisynthetic process was developed by PharmaMar, starting from the antibiotic safracin B. Since 2007, Trabectedin has been used in clinical practice for treatment of sarcomas, ovarian cancer and other malignant tumors.
Recently, it was found that this ascidian contains a consortium of various symbiotic bacteria with the main species being the γ-proteobacterium Candidatus Endoecteinascidia frumentensis. The use of Meta-Omics technology allowed characterizing this consortium and identifying 25 genes that catalyze formation of ET-743. Subsequent sequencing showed that the true producer of the active substance of Trabectedin is this symbiotic bacterium [33].
Actinobacteria are the most prevalent among the invertebrate symbiont bacteria. They appear to be involved in biosyntheses of many metabolites from marine invertebrates, including those that have been used as active substances in medicinal drugs [34]. Other symbiotic bacteria have also been found in marine invertebrates. For example, another large group of symbiotic bacteria, is present in sponges that participates in biosynthesis of various secondary metabolites. This group belongs to a higher-rank taxon – representative of a novel candidate phylum Poribacteria [35]. In addition, symbionts of marine invertebrate have been also identified as cyanobacteria, firmicutes, and microscopic fungi.
Cyanobacteria are known for their extremely active peptides, including dolastatins, the most potent from all the natural and synthetic compounds known to be toxic against tumor cells. They inhibit growth of tumor cells in picomolar concentrations. Dolastatins were first isolated from the mollusk Dolabella auricularia, but their genuine producers proved to be the cyanobacteria Symploca hydnoides and Lyngbya majuscula. A derivative of dolastatin 10 (13), the so-called monomethyl auristatin E (14) (Fig. 1), was selected as an active substance for designing of the anticancer drug Vedotin (Brentuximab or AdcetrisR) – representative of a new generation of targeted anticancer drugs. This is a conjugate of monoclonal antibodies with above-mentioned derivative of powerful marine-derived cytotoxin. Vedotin has been used since 2011 for treatment of lymphomas and other oncological diseases [36, 37], manufacturer – “BSP Pharmaceutical S.r.l.”, Italy.
MARINE ANTIBIOTICS WITH ANTIBACTERIAL PROPERTIES
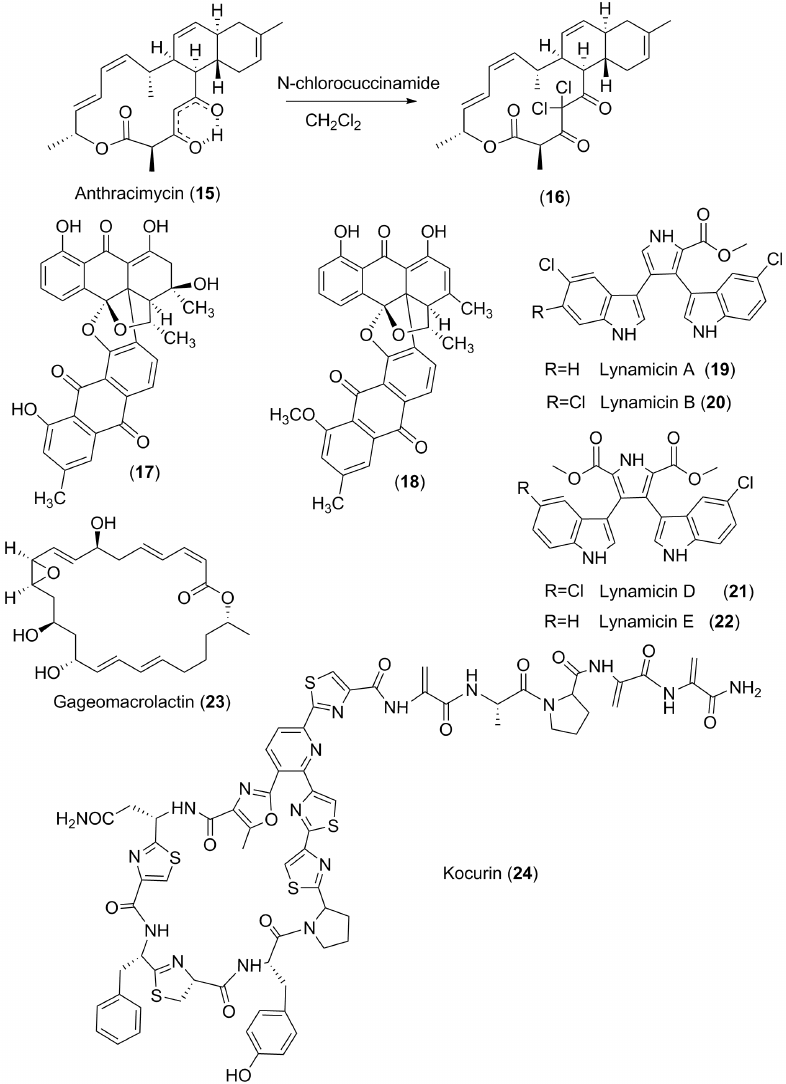
However, majority of the marine-derived antibiotics demonstrate strong antibacterial effect. The search for and discovery of these natural products are of great importance, since infectious diseases remain one of the main causes of death. Among them, there are quite different compounds in chemical structures: heterocyclic derivatives, quinoids, products of polyketide origin, peptides, etc. Dozens of research teams from many countries are actively searching for such substances. The laboratory of Professor William Fenical from the Scripps Institute of Oceanography (San Diego, USA), discovered several other marine antibiotics in addition to salinosporamides, for example anthracimycin (15) with a strong inhibitory effect against pathogenic bacteria. This antibiotic, isolated from the actinobacterium Streptomyces sp., strain T676, collected off the St. Johns Island near Singapore, inhibits Bacillus anthracis, the causative agent of anthrax, which causes a deadly disease and has already been used by terrorists who sent letters with spores of this bacterium to their targets. However, for the first time anthracimycin was obtained from another marine streptomycete, strain CNH365, collected from the bottom sediments off the coast of Santa Barbara, California, USA [38]. Chemical structure of the compound 15 was established by careful analysis of its NMR spectra and confirmed by X-ray diffraction analysis. Interestingly, its structure has an unusual hydrogen bond formed by two ketone groups with one of them in enol form (Fig. 2). When treated with N-bromosuccinimide in dichloroethane, anthracimycin forms a dichloro derivative (16), which also shows high antibacterial activity, especially against methicillin-resistant E. coli and some other pathogenic bacteria.
Fig. 2. Some antibacterial metabolites of marine bacteria.
The anthracimycin biosynthesis was studied at the genetic level. The identified “atc” gene cluster responsible for its biosynthesis consists of 53253 base pairs. It includes the genes of type I polyketide synthase and acyltransferase, which catalyze formation of malonyl and methylmalonyl coenzymes A involved in the construction of the skeletal system of 15. Expression of this gene cluster in the bacterium Streptomyces coelicolor led to the production of anthracimycin in this bacterium. 10 biosynthetic modules participate in this biosynthesis, the decaline cyclic system is first formed under their action, and after it the biosynthesis is completed by a thioesterase that catalyzes closure of the macrocycle to form a lactone bond [39].
New antibacterial agents (17, 18) (Fig. 2) were found in the Streptomyces sp. marine isolate from cyanobacterial mat collected off the coast of Puerto Rico. These unusual bis-anthraquinones show strong inhibitory effects against strains of vancomycin-resistant Enterococcus faecium, methicillin- and tetracycline-resistant Staphylococcus aureus. In addition, their synthetic derivatives have been also studied. The most effective antibiotic (17) demonstrates 50% bactericidal concentrations (MIC50) of 0.11, 0.23, and 0.90 µg per ml against the above-mentioned dangerous bacterial pathogens [40].
The bacterial strain NPS12745 was isolated from the sediment samples collected from the bottom of Missoni Bay in San Diego, California. Based on the nucleotide sequence of its 16S RNA gene, this bacterium was identified as a new species belonging to the genus Marinispora. The extract of this microorganism inhibits growth of both Gram-positive and Gram-negative pathogenic bacteria. The structures of the obtained antibiotics were established as new bis-indolyl pyrrole derivatives called lynamicins A-E (19-22) (Fig. 2). All of them demonstrated strong antibacterial effect. Lynamicin B showed the highest activity, and it inhibited strains of pathogenic bacteria Streptococcus pneumonia, Haemophilus influenzae, and Staphylococcus aureus at concentrations of 2-4 µg/ml [41].
Gageomacrolactins such as gageomacrolactin (23) (Fig. 2) from the bacteria isolated from the bottom sediments on the Gageo reef in the waters of the Republic of Korea, showed high antibacterial activity against both Gram-positive (Staphylococcus aureus, Bacilus subtilis) and Gram-negative bacteria (E. coli, Salmonella typhi, Pseudomonas aeruginosa) [42].
Peptide antibiotics were quite often found in the different marine bacteria, such as actinobacteria and cyanobacteria. Screening of 44 marine actinomycetes from the family Microccocaceae, isolated from sponges collected off the coast of Florida (USA), led to the discovery of new producers of antibiotics. They were identified by genetic methods as representatives of the genera Kocuria and Micrococcus and contained the genes of non-ribosomal peptide synthase and polyketide synthase. Three obtained isolates produced the peptide antibiotic Kokurin (21) (Fig. 2) – a new member of the family of thiazole-containing cyclic peptides [43].
A compound with similar structure, called antibiotic PM181104 (Fig. 2), was isolated by the Indian chemists after cultivation of a bacterium belonging to the same genus isolated from the sponge Spirastrella inconstans var. digitate, collected in Palk Bay, India. This antibiotic inhibits various pathogenic bacteria, including antibiotic-resistant strains, at concentrations of approximately 1 nM. In in vivo experiments it provided 100% protection of animals against sepsis caused by the methicillin-resistant S. aureus and vancomycin-resistant enterococci when administered into mice at doses of 5 or 10 mg/kg, respectively [44].
The number of promising antibiotics from marine organisms is increasing year after year, which attracts attention of more and more research groups around the world.
ANTIFUNGAL MARINE ANTIBIOTICS
Pathogenic fungi that cause severe mycoses remain one of the main threats to the cancer patients with weakened immune systems as a result of chemotherapy, as well as to the patients with AIDS and other diseases that initiate immunodeficiency. Such patients are often infected with opportunistic fungi such as representatives of the genera Candida, Aspergillus, and Cryptococcus leading to serious consequences up to fatal outcomes. About 500 thousand people die from such infections every year, and economic impact is about 12 billion dollars. The search for natural antifungal compounds and development of drugs on their basis is an urgent task of modern science.
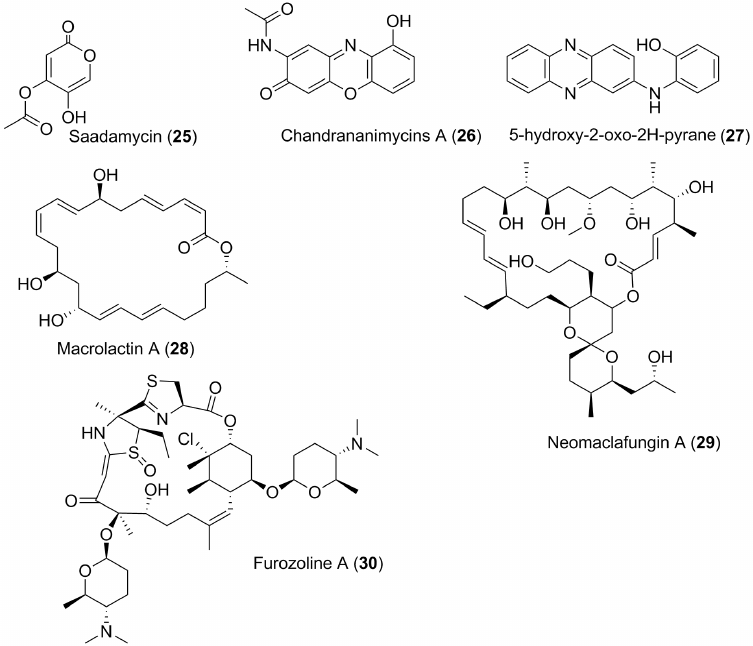
Some metabolites of marine bacteria, particularly actinomycetes, have become known for their antifungal properties in recent years. For example, saadamycin (25) (Fig. 3), a polyketide antibiotic from the actinobacterium Streptomyces sp. (a new species of this genus), isolated from the sponge Aplysina fistularis collected off the coast of Egypt, has a powerful antifungal effect against Candida albicans, Aspergillus spp. and Cryptococcus spp. with a minimum inhibitory concentrations (MIC) of 1-5.2 µg/ml and a minimum fungicidal concentrations (MFC) of 2-10 µg/ml [45]. Interestingly, scientists who discovered this new microbial source were able to increase production of the antibiotic 2.26-fold by treating its producer with ultraviolet rays. The resulting mutant strain Ah22 was then used to isolate and establish the structure of antibiotic itself. The structure of the compound 25 is not very complex. It is a product of polyketide biosynthesis that has an α-pyrone cycle, and proved to be a derivative of 5-hydroxy-2-oxo-2H-pyrane.
Fig. 3. Some antifungal metabolites from marine microorganisms.
A phenoxazine derivative, chandrananimycin A (26) (Fig. 3), and related compounds were isolated from the actinobacterium Actinomadura sp. collected from the bottom sediments of the Bay of Guangzhou (China). Compound 26 inhibits a number of fungi, including Mucor miehei, whose enzymes are used in cheese production [46].
A series of actinomycetes, including the BM-17 strain, were isolated from a sample of bottom sediments collected in the Arctic ocean. This strain has been identified as Nocardia dassonvilei. A new secondary metabolite (27) (Fig. 3) along with previously known phenazine antibiotics, was obtained when this strain was cultured. This compound shows significant antifungal activity against Candida albicans with MIC50 of 64 µg/ml and potent cytotoxic properties against a number of tumor cell lines [47].
Other marine macrolides also often show antifungal properties. For example, macrolactin A (28) (Fig. 3) and similar macrolactins isolated from the bacterium Bacillus subtilis collected in the area of the above-mentioned Gageo reef exhibit strong antifungal action against Aspergillus niger, Candida albicans, Botrytis cinerea, Colletotrichum acutum. Minimum inhibitory concentrations were as low as 0.04-0.3 µM [48]. Some related macrolides also exhibit potent antifungal effects [49].
Neomaclafungin A (29) (Fig. 3) from the bacterium Actinoalloteichus sp. NPS702, isolated from the sample of bottom sediment collected in Usa Bay (Japan) [50], shows significant antifungal activity against Trichophyton mentagraphites (MIC 1-3 µg/ml). After synthesis, its structure was revised [51].
Like bacteria, pathogenic fungi also develop drug resistance mechanisms and form fluconazole-and amphotericin-resistant strains. In particular, this is typical for the species belonging to the genus Candida. Candidoses affect about 400 thousand people annually. At the same time, fungi use various biochemical mechanisms to avoid lethal effects of the fungicides; they could use special transport systems to expel these drugs from the cells. Recently, metabolites active against such strains have been foundin marine organisms. For example, a polyketide metabolite furosoline A (30) was isolated from the bacterium Actinimadura sp. using liquid chromatography – mass spectrometry. This bacterium was isolated from the ascidian Ecteinascidia turbinata. Furozoline A (30) (Fig. 4) inhibits pathogenic fungi at a dose of 16 µg/ml. Successful use of 30 for treatment of mice with experimental multiple candidosis was demonstrated [52].
Fig. 4. Some marine antibiotics with antiviral properties.
It is of interest that some marine-derived steroids, such as polar steroids from the sponge Dysidea arenaria, increase activity of the antifungal drugs and cancel resistance of pathogenic fungi to fluconazole. At the same time, activity of the antifungal drugs after the treatment with these steroids can be increased 35-fold [53].
ANTIBIOTICS WITH ANTIVIRAL PROPERTIES
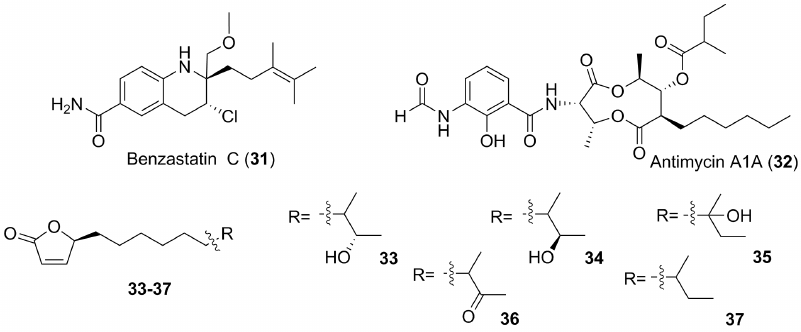
Only a few antiviral metabolites were found among the compounds isolated from marine actinobacteria. This may be due to a less intensive search for such substances compared to anti-tumor and anti-fungal natural marine-derived products. Benzastatin C (31) (Fig. 4) was obtained from the marine bacterium Streptomyces nitrosporeus. It demonstrates a dose-dependent inhibitory effects with EC50 1.92 and 0.53 µg/ml against HSV-1 and HSV-2 herpes viruses, respectively. Thus, it was more active in these tests than Ara C – a well-known drug that was also created on the basis of marine natural compounds and became the progenitor of the nucleoside-based antiviral drugs. At the same time, cytotoxic properties of 31 were low. Compound 31 shows also antiviral action against the vesicular stomatitis virus. It was shown that the presence of chlorine atom is necessary for the antiviral effect of this leader compound [54].
Antimycin 1A (32) (Fig. 4) [55] was isolated from the new species, Streptomyces kaviengensis, obtained from the sample of bottom sediment collected near New Ireland (Papua New Guinea). The isolated compound shows an extremely high antiviral effect (IC50 4 nM) against equine encephalitis virus. Its mechanism of action is related to the ability of inhibiting the electron transport network in mitochondria and suppressing de novo pyrimidine biosynthesis. Administration of the drug in vivo increases survival of the animals infected with this virus. Antimycin inhibits also other RNA viruses.
Adenovirus infections are well known to be associated with high mortality in patients with immunodeficiency. It is commonly recognized that there are no effective medical means to combat such infections. An actinobacterium belonging to the Streptomyces genus was isolated from one of fjords off the coast of Norway. It produced simple substances, butenolides, as mixtures of close related metabolites. Butenolide derivatives (33-37) (Fig. 4), isolated from the extracts of this streptomycete were active against adenoviruses. The most active compound shows an effective EC50 concentration of 91 µM and no noticeable cytotoxicity against human cells. These substances are of interest as model compounds for creating appropriate drugs [56].
ANTIPARASITIC COMPOUNDS FROM MARINE BACTERIA
For centuries, malaria claimed thousands of lives each year until first and second-generations of antimalarial drugs were developed. Quinine, chloroquine, and other therapeutics protected many millions of people, but unfortunately, later some forms of malaria resistant to such drugs emerged. As it is well known, this disease is caused by the parasitic protozoan, malarial plasmodium (Plasmodium falciparum), which is transferred to people via mosquito bites. In Africa, malaria is the most common disease; increasing annual numbers of malaria cases are detected in South America and South-East Asia. The search for new antimalarial compounds and development of appropriate drugs are becoming more and more important year by year. Recent successes in this scientific field was recognized by awarding the 2015 Nobel prize for creation of a new antimalarial drug artemisinin and new antiparasitic drug ivermectin developed on the basis of the corresponding natural compounds.
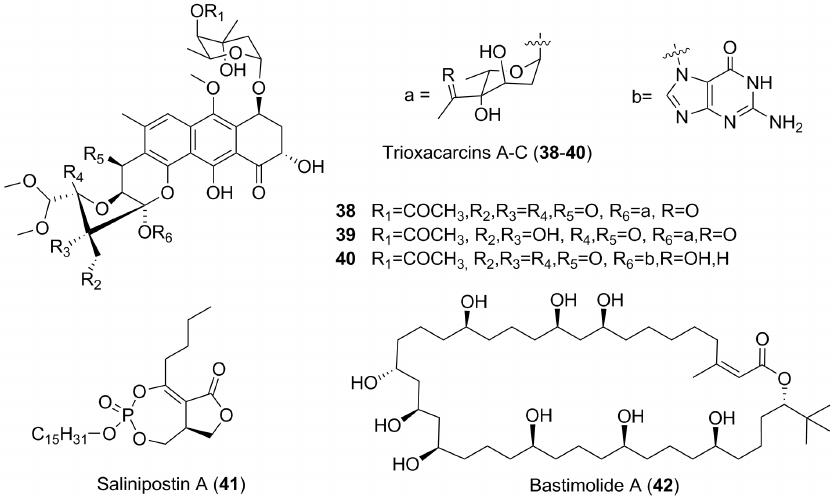
Marine-derived antimalarial compounds include, for example, trioxacarcins (38-40) (Fig. 5), produced by Streptomyses ochraceus and S. bottropensis. All the trioxacarcins show high antibacterial, antitumor, and some of them, particularly 38, antimalarial activities [57]. Salinipostins A-K inhibited the chloroquine-resistant W2 strains of P. falciparum with EC50 between 50 nM and 50 µM. Of these, salinipostin A shows the strongest effect (EC50 ~ 50 nM). It is expected that it would be more difficult to develop resistance to this compound because this drug candidate does not affected heme [58]. Salinipostins were obtained from the microbial strain of Salinispora sp. collected from the bottom of the Keawekaheka Bay, Hawaii. Salinipostin A (41) inhibits one of the growth stages of plasmodium.
Fig. 5. Some new antimalarial compounds from marine bacteria.
A new polyhydroxymacrolide, bastimolide A (42) (Fig. 5), was isolated from the tropical cyanobacterium Okeania hirsuta. It inhibited four drug-resistant strains of malaria plasmodium in nanomolar concentrations and was less toxic to both normal and tumor cells [59].
High antimalarial activity was also found for the above-mentioned salinosporamide A (1), which has been clinically studied as an antitumor drug. It is assumed that it acts on the 20S proteasome of the parasite. Administration of salinosporamide A at a dose of 130 µg/kg protects mice from malaria infection.
CONCLUSION
Recently, marine-derived natural compounds with antibiotic action have attracted more and more attention. In our country, these compounds are studied by not so many scientific groups, all these groups are concentrated only in two cities, Moscow and Vladivostok. Unexpectedly wide structural diversity of the metabolites from marine bacteria isolates and their promising biological effects makes it quite possible to develop new medical drugs based on them, and several such compounds have already become the active substances of new drugs or are currently undergoing preclinical or clinical trials. Structural novelty and rather complex chemical structure of many of them make it difficult to determine exact structures of such substances, in some cases this task can be solved only with the use of X-ray diffraction analysis. Unlike in the United States and Japan, in Russia targeted total asymmetric synthesis of natural compounds is not developing so well, therefore, additional attention should be given to this scientific direction. Sometimes, it is difficult to obtain the target substances in sufficient quantities for their in-depth study. The problem of obtaining of target substances is solved mainly using one of the two strategies: by creating microbial super producers with selection methods and mutagenesis or by transferring the corresponding gene clusters to easily cultured bacterial species.
Despite the existing difficulties and problems, we have several advantages in Russia when working in this area. First of all, these advantages include: (i) an extended coastline of the country and wide opportunities to collect the necessary biological materials, and (ii) the existence in Vladivostok of the collection of live marine bacteria and fungi, which contains about 4 thousand axenic strains collected in various geographical areas of the World. Nature has created unique structural resources in the marine environment, the study and use of which is an important task of basic research and applied science.
Funding. The work was financially supported by the Agreement No. 9188107-OK-20 for implementation of research work for the needs of the Pirogov Federal State Medical University, Russian Ministry of Health in 2020.
Ethics declarations. The authors declare no conflict of interest in financial or any other sphere. This article does not contain any studies with human participants or animals performed by any of the authors.
REFERENCES
1.URL: https://www.who.int/ru/news-room/fact-sheets/detail/tuberculosis.
2.Pendleton, J. N, Gorman, S. P., and Gilmore, B. F.
(2013) Clinical relevance of the ESKAPE pathogens, Exp. Rev. Anti
Infect. Ther., 11, 297-308, doi: 10.1586/eri.13.12.
3.URL: https://www.who.int/ru/news-room/detail/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed.
4.Schinke, C., Martins, T., Queiroz, S. C. N., Melo,
I. S., and Reyes, F. G. R. (2017) Antibacterial compounds from marine
bacteria, 2010-2015, J. Nat. Prod., 80, 1215-1228, doi:
10.1021/acs.jnatprod.6b00235.
5.Newman, D. J., and Cragg, D. J. (2007) Natural
products as sources of new drugs over the last 25 years, J. Nat.
Prod., 70, 461-477, doi: 10.1021/np068054v.
6.Jensen, P. R., and Fenical, W. (1996) Marine
bacterial diversity as a resource for novel microbial products, J.
Ind. Microbiology, 17, 346-351, doi: 10.1007/BF01574765.
7.Jensen, P. R., and Fenical, W. (1994) Strategies
for the discovery of secondary metabolites from marine bacteria:
ecological perspectives, Annu. Rev. Microbiol.,
48, 559-584, doi: 10.1146/annurev.mi.48.100194.003015.
8.Davidson, B. (1995) New dimensions in natural
products research: cultured marine microorganisms, Curr. Opin.
Biotechnol., 6, 284-291, doi:
10.1016/0958-1669(95)80049-2.
9.Pietra, F. (1997) Secondary metabolites from marine
microorganisms: bacteria, protozoa, algae and fungi. Achievements and
prospects, Nat. Prod. Rep., 14, 453-464, doi:
10.1039/NP9971400453.
10.Andryukov, B. G., Mikhailov, V. V., Besednova, N.
N., Bynina, T. S., and Matosova, E. V. (2018) The bacteriocynogenic
potential of marine microorganisms, Russ. J. Mar. Biol.,
44, 433-441, doi: 10.1134/S1063074018060020.
11.Eom, S. H., Kim, Y. M., and Kim, S. K. (2013)
Marine bacteria: potential sources for compounds to overcome antibiotic
resistance, Appl. Microbiol. Biotechnol., 97, 4763-4773,
doi: 10.1007/s00253-013-4905-y.
12.Rahman, H., Austin, B., Mitchell, W. J., Morris,
P S., Jamieson, D. J., et al. (2010) Novel anti-infective compounds
from marine bacteria, Mar. Drugs, 8, 498-518, doi:
10.3390/md8030498.
13.Mikhailov, V. V. (2005) Marine microbiology at
PIBOC FEB RAS, Herald of the Far Eastern Branch Russian Academy of
Science, 2, 145-152.
14.Romanenko, L. A., Uchino, M., Frolova, G. M.,
Tanaka, N., Kalinovskaya, N. I., Latyshev, N., and Mikhailov, V. V.
(2007) Sphingomonas molluscorum sp. nov., a novel marine isolate
with antimicrobial activity, Int. J. Syst. Evol. Microbiol.,
57, 358-363, doi: 10.1099/ijs.0.64441-0.
15.Manivasagan, P., Venkatesan, S., Sivakumar, S.,
and Kim, S. K. (2014) Pharmaceutically active secondary metabolites of
marine actinobacteria, Microbiol. Res., 169, 262-278,
doi: 10.1016/j.micres.2013.07.014.
16.Ul Hassan, S. S., Anjum, K., Abbas, S. Q.,
Akhter, N., Shagufta, B. I., Shah, S. A. A., and Tasneem, U. (2017)
Emerging biopharmaceuticals from marine actinobacteria, Environ.
Toxicol. Pharmacol., 49, 34-47, doi:
10.1016/j.etap.2016.11.015.
17.Weyland, H. (1969) Actinomycetes in North Sea and
Atlantic Ocean sediments, Nature, 223, 858, doi:
10.1038/223858a0.
18.Thomas, T. R. A., Kavlekar, D. P., and
LokaBharathi, P. A. (2010) Marine drugs from sponge-microbe association
– a review, Mar. Drugs, 8, 1417-1468, doi:
10.3390/md8041417.
19.Reddy, T. B. K., Thomas, A. D., Stamatis, D.,
Bertsch, J., Isbandi, M., et al. (2014) The genome online database
(GOLD) v.5: a metadata management system based on a four level
(meta)genome project classification, Nucleic Acids Res.,
43, 1099-1106, doi: 10.1093/nar/gku950.
20.Khan, S. T., Tamura, T., Takagi, M., and Shin-ya,
K. (2010) Streptomyces tateyamensis sp. nov., Streptomyces
marinus sp. nov. and Streptomyces haliclonae sp. nov.,
isolated from the marine sponge Haliclona sp., Int. J. Syst.
Evol. Microbiol., 60, 2775-2779, doi:
10.1099/ijs.0.019869-0.
21.Tian, X. P., Tang, S. K., Dong, J. D., Zhang, Y.
Q., Xu, L. H., Zhang, S., and Li, W. J. (2009) Marinactinospora
thermotolerans gen. nov., sp. nov., a marine actinomycete isolated
from a sediment in the northern South China Sea, Int. J. Syst. Evol.
Microbiol., 59, 948-952, doi: 10.1099/ijs.0.005231-0.
22.Schumann, P., Cui, X., Stackebrandt, E.,
Kroppenstedt, R. M., Xu, L., and Jiang, C. (2004) Jonesia
quinghaiensis sp. nov., a new member of the suborder
Micrococcineae, Int. J. Syst. Evol. Microbiol., 54,
2181-2184, doi: 10.1099/ijs.0.63223-0.
23.Han, S. K., Nedashkovskaya, O. I., Mikhailov, V.
V., Kim, S. B., and Bae, K. S. (2003) Salinibacterium
amurskyense gen. nov., sp. nov., a novel genus of the family
Microbacteriaceae from the marine environment, Int. J. Syst. Evol.
Microbiol., 53, 2061-2066, doi: 10.1099/ijs.0.02627-0.
24.Tian, X. P., Zhi, X. Y., Qiu, Y. Q., Zhang, Y.
Q., Tang, S. K., Xu, L. H., Zhang, S., and Li, W. J. (2009)
Sciscionella marina gen. nov., sp. nov., a marine actinomycete
isolated from a sediment in the northern South China Sea, Int. J.
Syst. Evol. Microbiol., 59, 222-228, doi:
10.1099/ijs.0.001982-0.
25.Maldonado, L. A., Fenical, W., Jensen, P. R.,
Kauffman, C. A., Mincer, T. J., Ward, A. C., Bull, A. T., and
Goodfellow, M. (2005) Salinispora arenicola gen. nov., sp. nov.
and Salinispora tropica sp. nov. obligate marine actinomycetes
belonging to the family Micromonosporaceae, Int. J. Syst. Evol.
Microbiol., 55, 1759-1766, doi: 10.1099/ijs.0.63625-0.
26.Jensen, P. R., Moore, B. S., and Fenical, W.
(2015) The marine actinomycete genus Salinispora: a model
organism for secondary metabolite discovery, Nat. Prod. Rep.,
32, 738-751, doi: 10.1039/c4np00167b.
27.Feling, R. H., Buchanan, G. O., Mincer, T. J.,
Kauffman, C. A., Jensen, P. R., and Fenical, W. (2003) Salinosporamide
A: A highly cytotoxic proteasome inhibitor from a novel microbial
source, a marine bacterium of the new genus Salinispora,
Angew. Chem. Int. Ed., 42, 355-357, doi:
10.1002/anie.200390115.
28.Williams, P. G., Buchanan, G. O., Feling, R. H.,
Kauffman, C. A., Jensen, P. R., and Fenical, W. (2005) New cytotoxic
salinosporamides from the marine actinomycete Salinispora
tropica, J. Org. Chem., 70, 6196-6203, doi:
10.1021/jo050511+.
29.Macherla, V. R., Mitchell, S. S., Manam, R. R.,
Reed, K. A., Chao, T. H., et al. (2005) Structure-activity relationship
studies of salinosporamide A (NPI-0052), a novel marine derived
proteasome inhibitor, J. Med. Chem., 48, 3684-3687, doi:
10.1021/jm048995+.
30.Miller, E. D., Kauffman, C. A., Jensen, P. R.,
and Fenical, W. (2007) Piperazimycins: cytotoxic hexadepsipeptides from
a marine-derived bacterium of the genus Streptomyces, J. Org.
Chem., 72, 323-330, doi: 10.1021/jo061064g.
31.Kalinovskaya, N. I., Romanenko, L. A.,
Kalinovsky, A. I., Ermakova, S. P., Dmitrenok, P. S., and Afiyatullov,
S. S. (2017) The antitumor antibiotics complex of aureolic acids from
the marine sediment-associated strain of Streptomyces sp. KMM
9048, Nat. Prod. Comm., 12, 571-577.
32.Rinehart, K. L. (1999) Antitumor compounds from
tunicates, Med. Res. Revs., 20, 1-27, doi:
10.1002/(SICI)1098-1128(200001)20:1<1::AID-MED1>3.0.CO;2-A.
33.Rath, C. M., Janto, B., Earl, J., Ahmed, A., Hu,
F. Z., et al. (2011) Meta-omic characterization of the marine
invertebrate microbial consortium that produces the chemotherapeutic
natural product ET-743, ACS Chem. Biol., 6, 1244-1256,
doi: 10.1021/cb200244t.
34.Blockley, A., Elliott, D. R., Roberts, A. P., and
Sweet, M. (2017) Symbiotic microbes from marine invertebrates: driving
a new era of natural product drug discovery, Diversity,
9, 49, doi: 10.3390/d9040049.
35.Bibi, F., Faheem, M., Azhar, E. I., Yasir, M.,
Alvi, S. A., Kamal, M. A., Ullah, I., and Naseer, M. I. (2017) Bacteria
from marine sponges: a source of new drugs, Curr. Drug Metab.,
18, 11-18, doi: 10.2174/13892002176661610130906 10.
36.Francisco, J. A., Cerveny, C. G., Meyer, D. L.,
Mixan, B. J., Klussman, K., et al. (2003) cAC10-vcMMAE, an
anti-CD30–monomethyl auristatin E conjugate with potent and
selective antitumor activity, Blood, 102, 1458-1465, doi:
10.1182/blood-2003-01-0039.
37.Zhou, Q., and Kim, J. (2015) Advances in the
development of site-specific antibody-drug conjugation, Anticancer
Agents Med. Chem., 15, 828-836, doi:
10.2174/1871520615666150302125448.
38.Jang, K. H., Nam, S. J., Locke, J. B., Kauffman,
C. A., Beatty, D. S., Paul, L. A., and Fenical, W. (2013)
Anthracimycin, a potent anthrax antibiotic from a marine-derived
actinomycete, Angew. Chem. Int. Ed., 52, 7822-7824, doi:
10.1002/anie.201302749.
39.Alt, S., and Wilkinson, B. (2015) Biosynthesis of
the novel macrolide antibiotic anthracimycin, ACS Chem. Biol.,
10, 2468-2479, doi: 10.1021/acschembio.5b00525.
40.Socha, A. M., LaPlante, K. L., and Rowley, D. C.
(2006) New bisanthraquinone antibiotics and semi-synthetic derivatives
with potent activity against clinical Staphylococcus aureus and
Enterococcus faecium isolates, Bioorg. Med. Chem.,
14, 8446-8454, doi: 10.1016/j.bmc.2006.08.038.
41.McArthur, K. A., Mitchell, S. S., Tsueng, G.,
Rheingold, A., White, D. J., Grodberg, J., Lam, K. S, and Potts, B. C.
M. (2008) Lynamicins A-E, chlorinated bisindole pyrrole antibiotics
from a novel marine actinomycete, J. Nat. Prod., 71,
1732-1737, doi: 10.1021/np800286d.
42.Tareq, F. S., Kim, J. H., Lee, M. A., Lee, H. S.,
Lee, J. S., Lee, Y. J., and Shin, H. J. (2013) Antimicrobial
gageomacrolactins characterized from the fermentation of the
marine-derived bacterium Bacillus subtilis under optimum growth
conditions, J. Agricult. Food Chem., 61, 3428-3434, doi:
10.1021/jf4009229.
43.Palomo, S., González, I., de la Cruz, M.,
Martín, J., Tormo, J. R., et al. (2013) Sponge-derived
Kocuria and Micrococcus spp. as sources of the new
thiazolyl peptide antibiotic kocurin, Mar. Drugs, 11,
1071-1086, doi: 10.3390/md11041071.
44.Mahajan, G., Thomas, B., Parab, R.,
Patel, Z. E, Kuldharan, S., et al. (2013) In vitro and
in vivo activities of antibiotic PM181104, Antimicrob. Agents
Chemother., 57, 5315-5319, doi: 10.1128/AAC.01059-13.
45.El-Gendy, M. M. A., and El-Bondkly, A. M. A.
(2010) Production and genetic improvement of a novel antimycotic agent,
saadamycin, against dermatophytes and other clinical fungi from
endophytic Streptomyces sp. Hedaya48, J. Indust. Microbiol.
Biotech., 37, 831-841, doi: 10.1007/s10295-010-0729-2.
46.Maskey, R. P., Li, F. C., Qin, S., Fiebig, H. H.,
and Laatsch, H. (2003) Chandrananimycins A approximately C: production
of novel anticancer antibiotics from a marine Actinomadura sp.
isolate M048 by variation of medium composition and growth conditions,
J. Antibiot., 56, 622-629, doi:
10.7164/antibiotics.56.622.
47.Gao, X., Lu, Y., Xing, Y., Ma, Y., Lu, J., Bao,
W., Wang, Y., and Xi, T. (2012) A novel anticancer and antifungus
phenazine derivative from a marine actinomycete BM-17, Microbial.
Res., 167, 616-622, doi: 10.1016/j.micres.2012.02.008.
48.Nagao, T., Adachi, K., Sakai, M., Nishima, M.,
and Sano, H. (2001) Novel macrolactins as antibiotic lactones from a
marine bacterium, J. Antibiot., 54, 333-339, doi:
10.7164/antibiotics.54.333.
49.Karpinsky, T. M. (2019) Marine macrolides with
antibacterial and/or antifungal activity, Mar. Drugs, 17,
241, doi: 10.3390/md17040241.
50.Sato, S., Iwata, F., Yamada, S., and Katayama, M.
(2012) Neomaclafungins A–I: oligomycin-class macrolides from a
marine-derived actinomycete, J. Nat. Prod., 75,
1974-1982, doi: 10.1021/np300719g.
51.Zhu, S., and Wu, Y. (2017) Synthesis and
configuration of neomaclafungin A, Chem. Asian J., 12,
2211-2215, doi: 10.1002/asia.201700950.
52.Wyche, T. P., Piotrowsky, J. S., Hou, Y., Braun,
D., Deshpande, R., et al. (2014) Furozoline A: marine-derived
polyketide with antifungal in vivo efficacy, Angew. Chem.
Int. Ed., 53, 1-5, doi: 10.1002/anie.201405990.
53.Abdelmohsen, U. R., Balasubramanian, S.,
Oelschlaeger, T. A., Grkovic, T., Pham, N. B., Quinn, R. J., and
Hentschel, U. (2017) Potential of marine natural products against
drug-resistant, viral, and parasitic infections, Lancet Infect.
Dis., 17, e30-e41, doi: 10.1016/S1473-3099(16)30323-1.
54.Lee, J. G., Yoo, I. D., and Kim, W. G. (2007)
Differential antiviral activity of benzastatin C and its dechlorinated
derivative from Streptomyces nitrosporeus, Biol. Pharm
Bull., 30, 795-797, doi: 10.1248/bpb.30.795.
55.Raveh, A., Delecta, P. C., Dobry, C. J., Peng,
W., Schultz, P. J., Blakely, P. K., and Miller, D. J. (2013) Discovery
of potent broad spectrum antivirals derived from marine actinobacteria,
PLoS One, 8, e 82318, doi:
10.1371/journal.pone.0082318.
56.Strand, M., Carlsson, M., Uvell, H., Islam, K.,
Edlung, K., Cullman, I., et al. (2014) Isolation and characterization
of anti-adenoviral secondary metabolites from marine actinobacteria,
Mar. Drugs, 12, 799-821, doi:
10.3390/md12020799.
57.Maskey, R. P., Helmke, E., Kayser, O., Fiebig, H.
H., Maier, A., Busche, A., and Laatsch, H. (2004) Anti-cancer and
antibacterial trioxacarcins with high anti-malaria activity from a
marine Streptomycete and their absolute stereochemistry, J.
Antibiot., 57, 771-779, doi: 10.7164/antibiotics.57.771.
58.Schulze, C. J., Navarro, G., Ebert, D., DeRisi,
J., and Lenington, R. G. (2015) Salinipostins A-K, long-chain bicyclic
phosphotriesters as a potent and selective anti-malarial chemotype,
J. Org. Chem., 80, 1312-1320, doi: 10.1021/jo5024409.
59.Shao, C. L., Lenington, R. G., Balunas, M. J.,
Centeno, A., Boudreau, P., et al. (2015) Bastimolide A, a potent
antimalarial polyhydroxy macrolide from a marine cyanobacterium
Okeania hirsuta, J. Org. Chem., 80, 7849-7855,
doi: 10.1021/acs.joc.5b01264.