REVIEW: Characteristic Features of Protein Interaction with Single- and Double-Stranded RNA
Alexey D. Nikulina
Institute of Protein Research, Russian Academy of Sciences, 142290 Pushchino, Moscow Region, Russia
Received April 29, 2021; Revised June 6, 2021; Accepted June 6, 2021
The review discusses differences between the specific protein interactions with single- and double-stranded RNA molecules using the data on the structure of RNA–protein complexes. Proteins interacting with the single-stranded RNAs form contacts with RNA bases, which ensures recognition of specific nucleotide sequences. Formation of such contacts with the double-stranded RNAs is hindered, so that the proteins recognize unique conformations of the RNA spatial structure and interact mainly with the RNA sugar-phosphate backbone.
KEY WORDS: RNA–protein interaction, single-stranded RNA, double-stranded RNA, RNP domain, ribosomal proteinsDOI: 10.1134/S0006297921080125
Abbreviations: dsRBD, double-stranded RNA-binding domain; dsRNA, double-stranded RNA; RNP, ribonucleoprotein; ssRNA, single-stranded RNA.
INTRODUCTION
RNA-binding proteins participate in many essential cellular processes, such as translation regulation, RNA maturation, transport and localization, mRNA splicing and degradation, etc. Studying structural properties of the RNA elements recognized by proteins, as well as deciphering structure of the specific RNA–protein complexes facilitate elucidation of the mechanisms of these processes and, subsequently, provide means to control them. Investigation of the RNA–protein complexes has started at the Institute of Protein Research more than 30 years with studies of structural and functional features of the complexes of bacterial ribosomal proteins with fragments of ribosomal RNA [1-7].
The problem of classification of RNA–protein interactions has been repeatedly discussed in numerous publications, starting from the classic review by Draper [8] in1999 and continuing with the latest article on the RNA–protein complexes by Corley et al. [9]. RNA regions specifically recognized by proteins can be single- or double-stranded. In the single-stranded RNA (ssRNA) fragments, most nucleotides are not paired with the complementary bases and remain accessible for the interaction with proteins. In the double-stranded RNA (dsRNA) fragments, bases are involved in the complementary interactions between the nucleotides, so that the atoms of the paired bases become unavailable for the interaction with external partners. ssRNA fragments are, as a rule, functionally active RNA regions (e.g., stem-loop structures with 3-8-nucleotide loop). In addition to the accessible single-stranded fragments, RNA molecules contain multiple elements with an intricate spatial structures, such as three-way junctions (combination of three RNA helices), bulges, pseudoknots, and others. Since most nucleotides in such elements form pairs, these RNA regions are often called dsRNA. Diversity of the dsRNA organization has led to adaptation of the interacting proteins to RNA spatial elements and emergence of unique features in the proteins specifically binding various RNA structures.
STRUCTURAL MOTIFS IN THE ssRNA-BINDING PROTEINS
RNP domain proteins. Proteins with ribonucleoprotein (RNP) binding domain [RNA-binding domain (RBD) or RNA-recognition motif (RRM)] are the most common ssRNA-binding proteins. At least 0.5-1.0% of human genes contain nucleotide sequences coding for RNP domains [10]. The RNP domain was identified for the first time in the middle of 1980s by comparing the sequences of protein components of small nuclear RNPs (snRNPs) [11, 12]. The RNP domain contains two conserved motifs: RNP1 composed of 8 mostly aromatic or positively charged residues (K/R-G-F/Y-G/A-F/Y-I/L/V-X-F/Y) and RNP2 in the N-terminal fragment of the protein, composed of six amino acid residues (I/L/V-F/Y-I/AL/V-X-N-L) [13]. These two motifs are linked by a sequence of approx. 40 amino acid residues. Total length of the RNP domain is 80-90 a.a.; it is formed by a four-stranded antiparallel β-sheet and two helices located at the same side of the β-sheet. RNP1 is a part of the β3 strand, while RNP2 is a part of the β1 strand.
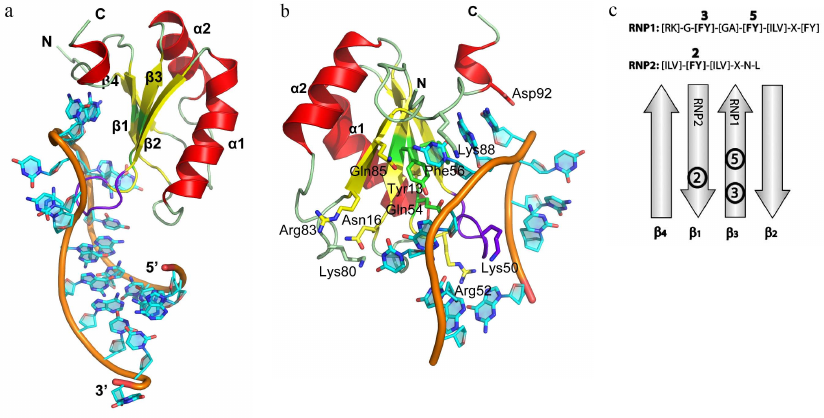
Resolving the structures of more than 30 complexes of RNP domain proteins (RNP proteins) with RNA fragments has allowed to analyze the details of interaction between these proteins and RNA and to elucidate the principles of recognition of the related RNA sequences [13-15]. A typical example of such proteins is U1A, which is a component of the snRNA U1 (one of the five snRNAs forming the spliceosome) [16, 17] (Fig. 1). RNP proteins can bind ssRNA regions of varying length – from 2 (CBP20 [18, 19], and nucleolin [20, 21]) to 8 nucleotides (U2B′′ [22]). In most cases, three conserved phenylalanine or tyrosine residues at positions 3 and 5 in the RNP1 and position 2 in the RNP2 (Fig. 1c) form contacts with two RNA bases, thus generating a continuous region of stacking interactions. Structural analysis has shown that, aside from few exceptions, RNP proteins retain conserved consensus motifs [13]. However, in all of them, the β-sheet plays the role of a “platform” that determines position of the bound RNA molecule, while the side chains of amino acids directed toward RNA act as “hooks,” providing protein–RNA interactions [23].
Fig. 1. Spatial organization of RNP proteins. a) Structure of the U1A protein in a complex with the snRNA II U1 hairpin (PDB 1URN). Elements of the protein secondary structure and 5′- and 3′-ends of the RNA fragment used in crystallization are indicated. Hereafter, the 3D structures were generated based on the atom coordinates using the PyMol program. b) Region of the U1A protein interaction with the U1 snRNA. Important amino acids involved in the interaction with the RNA bases via their side chains are shown. c) Location of RNP1 and RNP2 consensus sequences on the β-sheet of the RNA-binding domain. Aromatic amino acid residues of the consensus sequences are shown in bold.
An important feature of RNP proteins is the presence of several domains (the so-called tandem domains) for cooperative recognition of RNA molecules [24]. In this case, two RNP domains connected by a short linker interact with two consecutive RNA sequences. This structural organization allows to expand the RNA–protein interface area and to increase protein affinity toward ssRNA. Cooperative ssRNA recognition by the tandem RNP proteins was first observed for the Sxl protein, which specifically interacts with the UGU8 sequence [24]. The first nucleotides of the sequence (UGU) are recognized by the first RNP domain, while the following oligo(U) sequence is recognized by the second RNP domain. In this case, the fully unfolded ssRNA is recognized, which distinguished Sxl from the majority of RNP proteins that bind hairpin RNA structures with a short single-strand region.
Analysis of the ssRNA sequences revealed a relatively low selectivity of RNP proteins for them. For example, the RRM domain of the SRSF2 protein binds both the UCCAGU and UGGAGU sequences; moreover, the GG pair interacts with the same protein fragment on the β-sheet surface as the CC pair [24]. Therefore, RNP1 and RNP provide structural basis for the protein interaction with the single-stranded regions of RNA molecules, while specific RNA sequences are recognized by the protein terminal regions or by the partner proteins. It is commonly believed that RNP1 and RNP2 are the “LEGO blocks” for formation of the ssRNA-binding proteins functioning in tandem with other proteins [13, 15].
Specific features of RNP proteins can be found in the published reviews (e.g., [25]) and databases of classified protein families (Pfam PF00076, InterPro IPR000504).
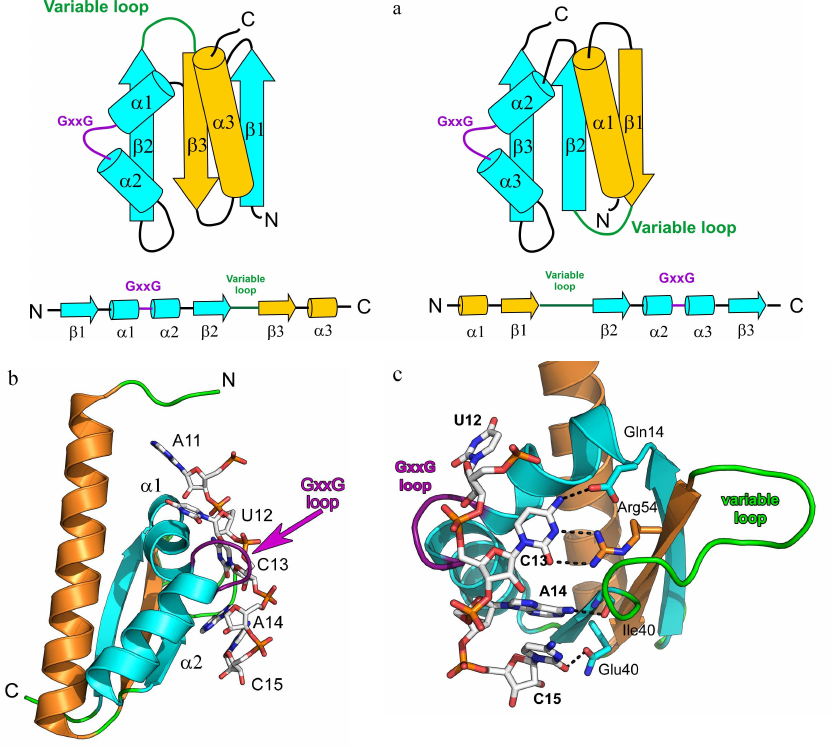
KH domain proteins. The second most common ssRNA-binding motif after the RNP domain is the KH domain [heterogeneous nuclear ribonucleoprotein K (hnRNP K) homology domain], which is approximately 70 a.a. long and contains the [ILV]-I-G-X-X-G-X-X-[ILV] motif in its central part [26]. The KH domain is a two-layer protein structure with a three-stranded β-sheet and three α-helices at its side that can form two different spatial structures. Eukaryotic (KH domain type I) and bacterial (KH domain type II) proteins share a common minimal core βααβ supplemented with one α-helix and one β-strand at the C-terminus (type I) or N-terminus (type II). Therefore, topology of the eukaryotic type I KH domain is βααββα with antiparallel β-sheet, while topology of the bacterial type II KH domain is αββααβ (Fig. 2a).
Fig. 2. Spatial organization of the KH domain proteins. a) Secondary structure elements in the type I (left panel) and type II (right panel) KH domains with the indicated protein core (cyan) and additional elements (yellow). b) Structure of the complex of Nova-2 KH3 protein (third domain of the mammalian Nova antigen from the family of RNA metabolism regulators in neurons) with the harpin RNA (PDB 1EC6); RNA fragment A11-C15 and GxxG motif of the protein loop are shown. c) Region of the Nova-2 KH3 protein interaction with the RNA tetranucleotide U12C13A14C15 (hydrogen bonds are shown with dashed lines).
Two α-helices of the protein central fragment are linked by the conserved GxxG loop in the KH domain motif. Proteins with the KH domain-like spatial structure, but lacking the GxxG motif, exhibit low affinity to RNA, but can modulate the RNA-binding activity of other proteins [27]. It is believed that the glycine residues of the GxxG loop are located very close to the atoms of the RNA sugar-phosphate backbone, and their substitution with other amino acids might create steric hindrances for the protein contacts with the RNA. Structural organization of the KH domain proteins ensure that four consecutive RNA bases are directed toward the protein surface and form a network of contacts with its amino acids (Fig. 2, b and c). This specifically recognized tetranucleotide usually contains pyrimidines at the first and fourth positions and adenine or cytosine at the second and third positions [28]. Cytosine at the second position is recognized due to formation of two hydrogen bonds between the O2 and N3 atoms of the base and the side chain of the arginine residue in the central β-strand. If the second position is occupied by adenine, the arginine in the protein is replaced with more mobile lysine residue. The base at the third position is specifically recognized via formation of two hydrogen bonds with the amide and carbonyl groups of the peptide backbone of one of the residues in the second (type I KH domain) or third (type II KH domain) β-strand. Both adenine and cytosine in this position can form two hydrogen bonds and are discriminated via formation of an additional hydrogen bond with the atom in the side chain of one of the α2-helix residues [29, 30].
It should be mentions that some KH domain proteins contain additional structural elements (e.g., signal transduction and activation of RNA fold (STAR) protein [31] or NusA protein with the tandem domains [32]) that promote protein affinity to RNA and decrease dissociation constant of the protein-RNA complexes from the micromolar to nanomolar range [33]. More detailed description of the structural properties of the KH domain proteins can be found in reviews [29, 34] and protein databases (Pfam PF00013, InterPro IPR004088).
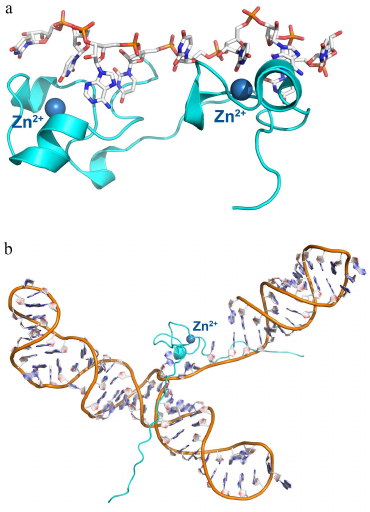
Protein with small repeated domains. Zinc finger proteins. Another interesting example of the RNA-recognizing motif is the zinc finger domain. Zinc finger domains were for the first time described as small (~30 a.a.) DNA-binding domains containing the [YF]-X-C-X-C-X(2,4)-C-X(3)-F-X(5)-L-X(2)-H-X(3,4)-H-X(5) consensus [35-37]. These proteins have the ββα topology, with two conserved pairs of histidine and cysteine residues binding a zinc ion that stabilizes the domain structure (Fig. 3). Zinc finger domains bind in the major grove of the double-stranded DNA helix, so that the atoms of the charged side chains form hydrogen bonds with the DNA bases. Zinc ion is not involved in the protein interaction with DNA; its main role is stabilization of the domain structure.
Fig. 3. Spatial organization of zinc finger proteins. a) Tis11d protein in a complex with the 5′-UUAUUUAUU-3′ RNA (PDB 1RGO). b) MMLV protein in a complex with a signaling RNA (PDB 1U6P).
Later, it was found that some RNA-binding proteins also contain zinc finger domains. Thus, the Tis11d protein, which regulates mRNA stability by binding to the class II AU-rich element (ARE) in the 3′-untranslated region (3′-UTR) of the target mRNA and promotes its deadenylation and degradation [38, 39], contains two zinc finger domains with C-X(8)-C-X(5)-C-X(3)-H (CCCH-type) motifs. The nucleocapsid MMLV (Moloney murine leukemia virus) protein contains one zinc finger domain (Fig. 3) [40, 41]. Each zinc finger domain of the Tis11d protein specifically recognized the single-stranded UAUU sequence in the class II AU-rich element (ARE) in the 3′-untranslated region (UTR) of the target and promotes its deadenylation and degradation. In this complex, four RNA bases are located in the pocket formed by each domain and form stacking interactions with the side chain of phenylalanine (from the loop between the third cysteine and the histidine residue of the zinc finger) and tyrosine (from the loop between the second and the third cysteines of the zinc finger). Selectivity of this interaction is provided by the contact between the atoms of the RNA bases and atoms of the zinc finger peptide backbone (with the only one exception, such as contact formation with the side chain of Glu157).
The MMLV protein is especially interesting, as it contains the minimal zinc finger domain (termed knuckle) of the C-X(2)-C-X(3)-H-X(4)-C (CCHC) type (Fig. 3b). Similar, to the Tis11d protein, MMLV protein interacts with RNA via stacking of bases between tyrosine and tryptophan residues of the zinc fingers, but in this case, the atoms of the bases form contacts mostly with the side chains of amino acids.
To summarize available data, zinc finger proteins recognize RNA due to formation of hydrogen bonds between amino acid residues and RNA, but the major role in these contacts belongs to the stacking interactions between the side chains of aromatic residues and RNA bases. The presence of several zinc finger motifs increases protein affinity and specificity toward RNA; however, these repeated domains exhibit no pronounced preference for DNA or RNA, which is a characteristic feature of the zinc finger proteins.
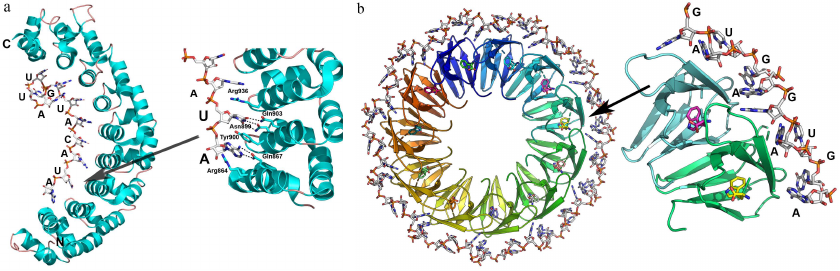
Proteins with the PUM domains (PUF repeats). Another example of small repeated domains interacting with the extended ssRNA tracts is the PUM-HD (Pumilio-homology domain) named after the Pumilio protein involved in the regulation of Drosophila maturation [42]. PUM-HD proteins bind to the 3′-UTRs in mRNAs, thus regulating expression of multiple genes [43, 44]. Human Pumilio1 PUM-HD protein contains eight PUF repeats of 37 a.a. each and the N- and C-terminal sequences structurally similar to the PUF repeats (Fig. 4a) [42].
Fig. 4. Structural organization of PUM domain proteins and TRAP. a) Human Pumilio1 PUM-HD protein in complex with the 10-nucleotide ssRNA (PDB 1M8Y). Right panel) details of protein–RNA interaction with the in the region of contact with one of the repeats. b) TRAP in a complex with the 53-nucleotide ssRNA containing 11 GAG triplets separated by the AU dinucleotides (shown surrounding the protein) (PDB 1C9S). Tryptophan molecules involved in the RNA binding are located at the surface of the protein monomer β-sheet. Right panel) detailed view of the RNA–protein contact area; (recognized GAG triplets and AU spacers are indicated).
Each PUF repeat consists of three α-helices that form a curved structure. ssRNA binds to the concave inner part of the protein, so that each base is located between the neighboring repeats, whereas the RNA sugar-phosphate backbone forms no contacts with the protein. Most nucleotides stack with the side chains of tyrosine and tryptophan residues, and atoms of the Watson–Crick base edge form hydrogen bonds with the side chains of polar and charged amino acids of the α2-helices of each repeat [42]. Interestingly, uracil binds to glutamine and asparagine, while adenine binds to glutamine and cysteine and guanine interacts with glutamine and serine. Substitution of these residues alters the protein specificity to the ssRNA sequences [42, 45, 46]. This is an extreme example of the principle of using of repeated domains for increasing protein affinity to RNA, as each domain represents a PUF repeat that recognizes an individual nucleotide.
TRAP. TRAP (tryptophan RNA binding attenuation protein) is another representative of the proteins with large number of small RNA-binding domains. In some bacilli, TRAP regulates expression of proteins involved in the synthesis of L-tryptophan. After binding free tryptophan molecules, TRAP interacts with the 5′-UTR of mRNA and forms the terminator loop, resulting in the transcription termination [47, 48].
TRAP is an entirely β-structure protein (two-layer antiparallel β-sandwich), 11 monomers of which form a symmetrical ring-shape quaternary complex (Fig. 4b). Tryptophan binds to TRAP between the two β-layers. In the protein complex with the 53-nt ssRNA, each of the 11 GAGAU repeats interacts with one of the protein monomers [47]. The GAG triplets are located between the two neighboring monomers and interact with them, while the AU dinucleotide acts as a spacer. Specific recognition of adenine (A) and third guanine (G) in each GAG triplet is ensured by a network of hydrogen bonds, whereas the RNA sugar-phosphate backbone forms no contacts with the protein atoms, except the hydrogen bond between the 2′-OH group of ribose and amide group of Phe32. Apparently, this contact is important for the selective binding of RNA, as affinity of TRAP to the DNA molecule with the same nucleotide sequence is 10,000 times lower [49].
Proteins with OB fold domain. The OB fold domain named due to its presence in the proteins capable of oligonucleotide/oligosaccharide binding represents a β-barrel composed of five β-strands. The OB fold domains have been found in multiple proteins with different functions and size (70-150 a.a.) [50]. According to the SCOP structural classification, the OB fold is subdivided into 16 superfamilies, only one of which is named “nucleic acid-binding proteins” (superfamily SCOP 50249). The OB fold does not determine the RNA-binding ability of the protein molecule, but rather serves as a platform for formation of proteins with such ability, as it represents a rigid and stable structure resulting from the tight packing of the β-sheet.
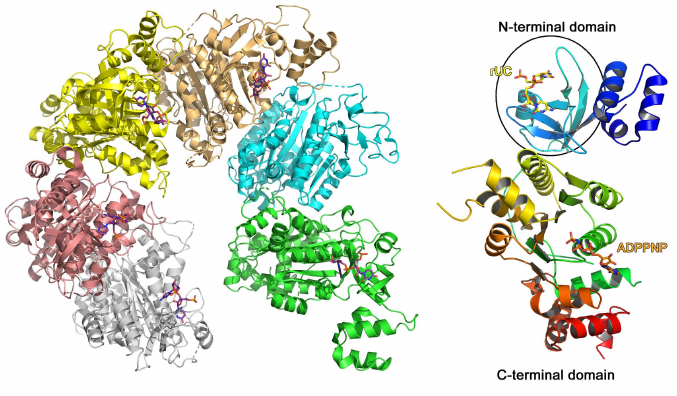
A typical example of the RNA-binding protein from this family is the Escherichia coli transcription termination factor Rho (Fig. 5) [51]. In solution, Rho monomer consists of two domains: smaller N-terminal RNA-binding domain and larger C-terminal ATPase domain. In addition to the β-barrel (OB fold), the N-terminal domain contains 47 a.a. that form three α-helices at the protein N-terminus. In the complex of this domain with oligo(C) RNA, two cytidines bound to the region of the β2- and β3-strands are covered with the β1-β2 loop and the α4-helix on one side and the β2-β3 loop on the other side [52]. The observed contacts between the protein amino acids and nucleotides suggests Rho protein specificity toward cytidines. No bonds are formed between the ribose 2′-OH group and protein, which is in agreement with the Rho ability to bind both RNA and DNA [52].
Fig. 5. Spatial structure of the E. coli hexameric transcription termination factor Rho in complex with short RNA and ADPPNP nucleotide (PDB 1PVO). Right panel) Rho monomer with bound UC dinucleotide and ADPPNP (OB fold domain is shown in the oval).
Other OB fold domain proteins can bind both RNA and DNA, for example, cold shock domain (CSD) proteins (Pfam PF00313, InterPro IPR002059) [53-55], CSD domain of the eukaryotic YB-1 protein [56], domains of bacterial S1 ribosomal proteins (InterPro IPR000110), etc. OB folds are structurally homologous to Sm folds (see below).
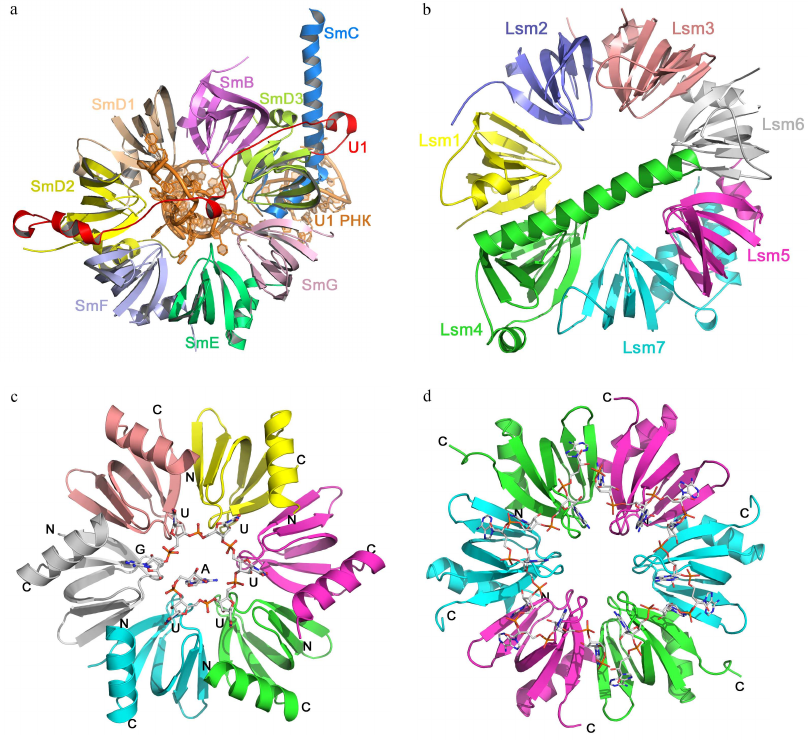
Sm fold proteins. The Sm fold was named after the Sm proteins, components of eukaryotic small nuclear RNPs (snRNPs) [57]. Seven homologous Sm proteins (B/B′, D3, D2, D1, E, F, and G) form the conserved part of the spliceosome snRNP with several uridine-rich snRNAs [58-60]. Sm proteins consist of approx. 80 a.a. organized in a five-stranded β-barrel with N-terminal α-helix structurally homologous to the OB fold (Fig. 6). In the snRNP, Sm proteins form a ring-like heteroheptamer (similar to the Rho protein), but the ring is fully closed and contacts between the neighboring protein monomers are formed by the outer β-strands, and not by the additional C-terminal domains. It should be mentioned that the Sm proteins form heptamers only in the presence of snRNAs [58]. Eukaryotes besides contain the Lsm (like-Sm) proteins homologous to the Sm proteins [61]. They are components of RNPs involved in the maturation and processing of mRNAs and tRNAs, mRNA decapping, and other processes [62]. Lsm proteins can also oligomerize into heteroheptamers, but this assembly does not require the presence of RNA [63].
Fig. 6. Structural organization of Sm/Lsm-proteins. a) Human minimal U1 snRNP. U1 snRNA is in the center of the heptamer; Sm proteins are shown with different colors. b) Yeast heteroheptameric Lsm1-7 complex (PDB 4C92). c) Staphylococcus aureus Hfq complex with AU5G RNA (PDB 1KQ2). d) E. coli Hfq complex with oligo(A) RNA (PDB 3GIB)
Homologues of eukaryotic Sm/Lsm proteins have been found in archaea and bacteria. Bacterial Lsm proteins are called Hfq and act as global regulator of gene expression [64, 65]. Archaeal Lsm proteins were named SmAPs (Sm archaeal proteins); their function in the cells remains poorly understood [61, 65]. Both archaeal and bacterial Lsm proteins form stable ring-like oligomers (homoheptamers and homohexamers, respectively).
Sm/Lsm proteins bind oligo(U) RNA in the oligomer inner cavity from the side of the monomer α-helices (Fig. 6c) [66]. Each uridine is located in the pocket formed by two neighboring monomers, while uracils form stacks with the aromatic side chains of amino acid residues of the β2-β3 and β4-β5 loops. Atoms of the RNA bases form hydrogen bonds with the side chains of charged amino acids. Since the size of the inner cavity is small, not all protein monomers have equal contact with the bases, due to ssRNA has to “enter” and “exit” the complex.
Bacterial Hfq proteins also exhibit affinity toward oligo(A) RNA [67, 68], which binds to the Hfq hexamer at the side opposite to the site of oligo(U) RNA binding (Fig. 6d). Adenines located in the hydrophobic pocket stack with the aromatic side chains and form hydrogen bonds with the side chains of other amino acids. The distance between specifically bound nucleotides (adenines) is greater than in the case of oligo(U) RNA binding. Nucleotides located between them can interact with amino acid residues [67] or can be exposed at the protein surface without forming specific contacts with the protein atoms [69].
Sm/Lsm proteins illustrate the principle of domain repetition for increasing protein affinity toward RNA, more exactly, toward its particular bases (uridine or adenine). Organization of the secondary structure elements serves as a basis for generation of the stable multimeric complexes that act as a single structure, while the β-sheet surface is not utilized for the interaction with RNA. The regions involved in RNA recognition are located in the protein pockets formed by the loops that link the elements of the protein secondary structure. These regions include hydrophobic residues stacking with nucleotide bases, as well as charged/polar amino acids forming a network of hydrogen bonds with the atoms of the bases.
PROTEINS INTERACTING WITH DOUBLE-STRANDED RNAs
Double-stranded RNA-binding domain (dsRBD). A single conserved structure called the double-stranded RNA-binding domain (dsRBD) is recognized in the proteins interacting with dsRNA [70, 71]. It is the second most common domain in the RNA-binding proteins after the RNP domain responsible for ssRNA binding. It is a small domain (65-70 a.a.) composed of three-stranded antiparallel β-sheet with the N- and C-terminal helices located at one of its surfaces. dsRBD was first identified by comparing amino acid sequences of eukaryotic proteins with high specificity toward dsRNA, but low specificity to ssRNA and to any DNA. Substitutions in the nucleotide sequence of dsRNA had no effect on the affinity of dsRBD proteins toward it [72].
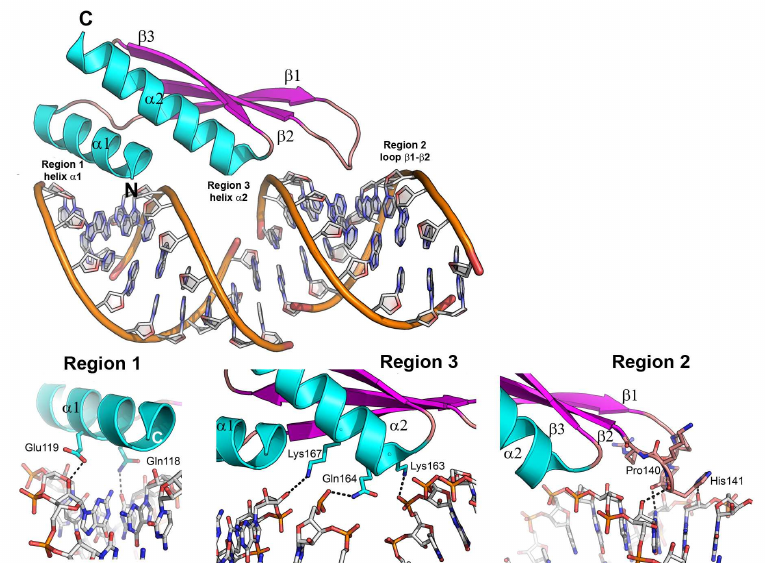
One of such proteins is the second domain of the RNA binding protein A from Xenopus laevis (Xlrbpa-2), which interacts with the 10-bp dsRNA (GGCGCGCGCC)2 via three regions (Fig. 7) [73]. The first (N-terminal α-helix) and the second (β1-β2 loop) regions form contacts with two fragments of the RNA small groove separated by a helix turn. The third region (C-terminal helix) interacts with a fragment of the RNA major groove. In all three regions, protein residues contact with the atoms of the RNA sugar-phosphate backbone, mostly, with the ribose 2′-OH group. Regular structure of the dsRNA A-form minor groove is distorted in the β1-β2 loop area (region 2); this distorted region contacts with the conserved histidine residue essential for the protein interaction with the dsRNA. Interestingly, the dsRNA binding to the protein depends not only on the amino acid residues forming direct contacts with the RNA molecule, but also on a number of hydrophobic residues that determine correct folding of α-helices and appropriate orientation of charged residues involved in the RNA binding [70].
Fig. 7. Structure of the Xlrbpa-2 complex with dsRNA (PDB 1DI2). Two neighboring dodecamer dsRNA fragments (GGCGCGCGCC), Xlrbpa-2 secondary structure elements, and regions of protein contacts with the dsDNA are shown. Bottom panel, proposed areas of RNA–protein contacts.
Analysis of the known structures of dsRBD proteins in complexes with dsRNAs shows that all these proteins form very few contacts with the RNA bases. Almost all protein interactions occur with the sugar-phosphate backbone atoms and ribose 2′-OH group, which is the most significant difference between the dsRBD proteins and ssRNA-recognizing proteins.
Additional secondary structure elements, such as an elongated N-terminal α-helix or repeated domains, can be used to increase affinity of the dsRBD proteins to dsRNAs [70, 71, 74]. Nevertheless, in any case, dsRBD proteins interact with the sugar-phosphate backbone atoms, and not with the atoms of bases in dsRNA.
Ribosomal proteins. Ribosomal proteins comprise an interesting example of proteins recognizing complex RNA spatial structures. Ribosome is a macromolecular complex composed of several ribosomal RNAs (rRNAs) and over 50 proteins. rRNA has an intricate spatial structure capable of self-organization in vitro. Ribosome assembly is a cooperative process that occurs via a strictly ordered series of events after being initiated by the interaction of ribosomal proteins with the specifically recognized rRNA sequences. Protein binding to the rRNA leads to compaction of the formed RNP particles and provides a platform for further association of other ribosomal proteins [75, 76]. The primary binding ribosomal proteins interact with rRNA independently on other ribosomal components. rRNA protein binding regions have a very complex spatial structure [77, 78]. The resulting complexes can be considered as protein-dsRNA complexes. It should be noted that some primary ribosomal proteins regulate their own synthesis by interacting with their own mRNA. Therefore, they can specifically recognize two different RNA targets. Autoregulation of ribosomal protein biosynthesis is a very interesting problem in the studies of principles of RNA–protein recognition and discrimination between two RNA molecules by the same protein. Below, we discuss three bacterial ribosomal proteins that are primary ribosomal protein and regulators of the bacterial operon translation. No such studies have been performed for eukaryotic proteins. It should be mentioned that the new nomenclature of the ribosomal proteins was introduced in 2014 to eliminate discrepancies in the naming of bacterial, archaeal, and eukaryotic proteins [79], according to which the universally conserved ribosomal proteins (i.e., found in the organisms from all three Kingdoms) were designated with prefix “u”, while the proteins typical for bacteria and eukaryotes only are designated with prefixes “b” and “e”, respectively.
Ribosomal protein uS8. The universally conserved ribosomal protein uS8 (hereafter, S8) is one of the ribosomal proteins forming the platform of the small ribosomal subunit [80-84]. It is essential for the correct assembly of the 16S rRNA central domain [81, 83, 85]. Mutations in S8 result in the defective association of ribosomal subparticles in the full-size ribosome [86]. S8 protein binds to the fragment of the h21 helix in the 16S rRNA in the region of nucleotides 595-598/640-644 [87-89]. S8 is also involved in the negative feedback regulation of transcription of the spc operon in E. coli [90, 91]. The structure of the protein binding region in the spc mRNA closely resembles that of the protein-binding sequence in the h21 helix of 16S rRNA, which initially gave rise to the idea of recognition based on the structurally similar elements of two RNAs [90, 92, 93].
The structure of the complex of S8 with a 16S rRNA fragment demonstrated that two protein domains interact with two neighboring fragments of the RNA minor groove, while the protein forms a “bridge” over the major groove (Fig. 8a) [4]. The majority of amino acid residues forming contacts with the rRNA are located in the protein C-terminal domain and interact mostly with the RNA sugar-phosphate backbone. Conserved nucleotides 595-598/640-644 are responsible for the distortions in the dsRNA A-form by forming two nucleotide triplets linked by hydrogen bonds. The same nucleotides form the S8-recognized interface on 16S rRNA [4].
The structure of E. coli S8 protein in complex with the fragment of the spc operon mRNA revealed slight differences in the sequence and spatial structure of the interacting regions in rRNA and mRNA [94]. The contacts between amino acids and mRNA are distributed in a manner similar to that observed in the ribosomal complex. Despite the similarity of RNA regions recognized by the S8 protein, S8 exhibits significantly different affinities toward rRNA and mRNA, reasons for which have not been explained yet.
Fig. 8. Structures of ribosomal protein complexes with RNA. a) S8 from Methanococcus jannaschii in complex with 16S rRNA fragment (PDB 1I6U); amnio acids residues forming contact with rRNA are indicated. b) S15 from Thermus thermophilus in complex with specific 16S rRNA fragment (PDB 1DK1). 16S rRNA helices h21, h22, and h23 and S15 protein α-helices are indicated. Left panel) view from the S15 protein side; right panel) the same image turned by 90° around the vertical axis.
Comparison of the S8 protein complexes with rRNA/mRNA and Xlrbpa-2 protein complex with dsRNA reveals obvious similarity between the structures. Indeed, the region of the dsRNA interaction with the protein in both cases is represented by a distorted regular structure of the RNA minor groove, where single amino acid residues interact with RNA bases. This region is surrounded by the areas of contacts formed by the side chains of polar and charged amnio acid residues with the RNA sugar-phosphate backbone, which shield the area of protein-RNA recognition from water molecules and ions and increase the energy gain upon the RNA–protein interaction. However, in the case of 16S rRNA, the distortions in the RNA helical structure are significant and result from the formation of two nucleotide triplets. In the case of Xlrbpa-2, this distortion is less pronounced, as it is caused by the bulging out of a single nucleotide. Therefore, despite the apparent similarity, spatial structures of the protein-binding regions in RNA differ significantly.
Ribosomal protein uS15. The universally conserved ribosomal protein uS15 (hereafter, S15) is involved in formation of the platform of the small ribosomal subunit. It is the first protein to attach to the 16S rRNA central domain thus mediating subsequent binding of the ribosomal proteins S6, S18, S11, and S21 [80, 81, 95, 96]. Similarly to the S8 protein, S15 regulates transcription of its own operon. When synthesized in excessive amounts, this protein binding to the corresponding mRNA and suppresses its own biosynthesis [97-100].
The ribosomal protein S15 is a small protein composed of four α-helices packed into a single domain. S15 binds 16S rRNA in the region of the h20, h21, and h22 helices (Fig. 8b) [83, 101]. Out of the two dozen amino acid residues interacting with rRNA, only four are positively charged, while most of the contacts are formed between the polar side chains of the residues and atoms of the rRNA sugar-phosphate backbone [5, 95]. The protein interacts with 16S rRNA in two regions: in the area of three-way junction between the 16S rRNA helices and the GU/GC motif of the ds-rRNA minor groove located one turn from the first region. An important feature of the first region is formation of the base triplet essential for fixation of the three-way junction. This triplet directly or via magnesium ion interacts with the side chains of amino acid residues from the α3-helix. The neighboring nucleotides interact with the residues of the α1-α2 loop. Most of these contacts are formed with the sugar-phosphatase backbone of the RNA minor groove.
The second region of the RNA–protein contacts is located in the upper part of the h22 helix, where the S15 protein specifically binds to the highly conserved nucleotides GU/GC of the minor groove. Conserved amino acid residues of the α2-α3 loop directed toward the RNA form hydrogen bonds directly or via the water molecule with all four listed nucleotides. Based on the structural and biochemical data, it was suggested that the region of conserved GU/GC nucleotides is specifically recognized by the protein residues, while protein interaction with the three-way junction in the 16S rRNA stabilizes their mutual position during the small subunit assembly [5].
Modeling of the structure of S15 protein complex with the specifically recognized mRNA fragment revealed that this fragment has the pseudoknot structure with an additional binding region [102-106]. The structures of the regions recognized in rRNA and mRNA molecules turned out to be somewhat similar, which explains the possibility of recognition of two different RNA types by the same protein. The authors termed this similarity of the spatial structure of recognized fragments in two different RNA molecules molecular mimicry [106]. However, difference in the structure of mRNA (pseudoknot) and rRNA (three-way junction) fragments might be the reason for different affinity of the S15 protein toward these RNA molecules.
Ribosomal protein uL1. The universally conserved ribosomal protein uL1 (hereafter, L1) together with the 23S rRNA helices H76, H77, and H78 forms a flexible functionally important fragment of the large ribosomal subunit called the L1 protuberance [107]. In E. coli cells, L1 protein regulates translation of the L11 operon that includes genes for the ribosomal proteins L1 and L11 [108, 109].
L1 consists of two domains: the first domain is formed by the N- and C-terminal parts of the protein with the second domain located between them [107]. The first domain has the two-layer split abc/d unit, or split β-α-β structure, and the second domain has the three-layer Rossmann fold unique for the ribosomal proteins. The two domains are connected by a flexible link and can change their relative orientation in solution [110].
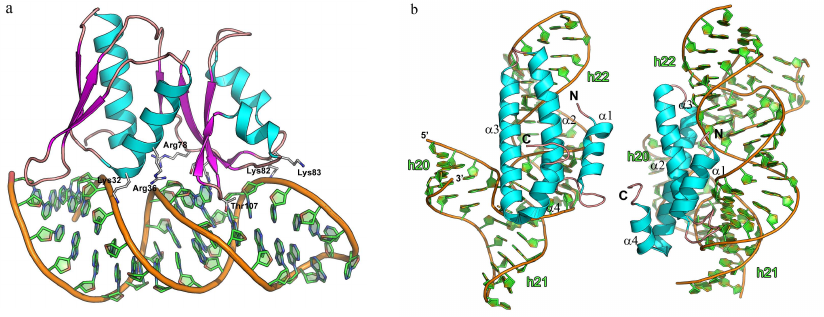
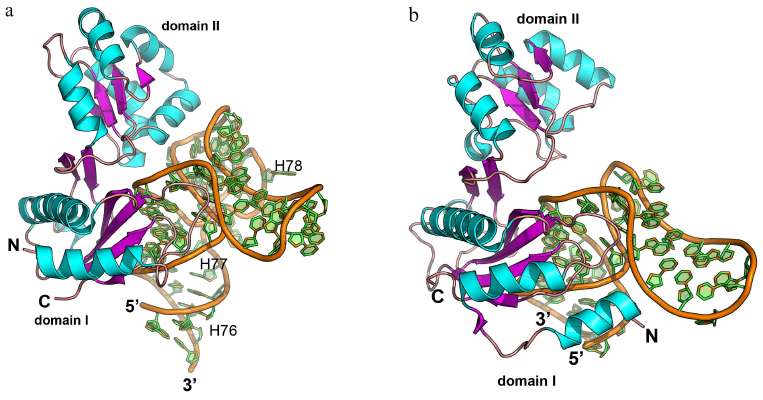
The complexes of L1 protein with the 23S rRNA fragments have two areas of contact (Fig. 9a) [3]. One of these areas is located at the surface of domain I. Three strands of the β-sheet form a slightly concave surface of contact with the 23S rRNA helix H77. Such protein architecture resembles the structure of the RNP domain, which also contains β-sheet surface involved in the interaction with RNA. However, sequence of the L1 protein does not include the RNP consensus motifs, and the L1 protein residues interact with the dsRNA minor groove (and not with the ssRNA). Most contacts between the protein and RNA in the region of the first domain are represented by the hydrogen bonds with the RNA sugar-phosphate backbone, although some hydrogen bonds are formed with the RNA bases.
Fig. 9. Structures of ribosomal L1 protein complexes with RNA. a) L1 from Sulfolobus solfataricus in complex with a fragment of 23S rRNA from T. thermophilus (PDB 1MZP). 23S rRNA helices H76, H77, and H78 are indicated. b) L1 from T. thermophilus in complex with a fragment of mRNA from Methanococcus vannielii (PDB 2HW8). The structures are oriented in the same manner relative to the L1 protein domain I.
The second region of the RNA–protein contact involves amino acid residues of the α4-helix and α5-β6 loop of the second domain, which form contacts with the 23S rRNA structure generated by two long RNA loops. Majority of the interactions are between the charged side chains of lysine and arginine residues and atoms of the RNA sugar-phosphate backbone. The number of amino acid residues interacting with RNA in this region is less than in the first region; therefore, it was suggested that the binding with 23S rRNA is determined by the amino acid residues of the first domain, while the contacts of the second domain stabilize the formed rRNA–protein complex.
Analysis of the spatial structure of the L1 protein complexes with the mRNA fragment revealed the reason for a significant difference in the protein affinity toward rRNA and mRNA [111-113]. Although mRNA fragments recognized by the L1 protein have similar nucleotide sequence and spatial structure, they lack one of the RNA loops, and therefore, the second RNA region involved in the contact with the protein is shortened (Fig. 9b). This results in almost complete absence of contacts in the second region and noticeable decrease in the L1 protein affinity toward mRNA. It was demonstrated later that the isolated L1 protein domain I exhibits lower affinity toward mRNA vs. rRNA and is involved in the discrimination between different types of RNA [114, 115].
CONCLUSIONS
Protein interaction with ssRNAs involves multiple RNA-recognizing motifs. An important role in such interaction in many cases belongs to the protein β-sheet that serves as a “platform” for the RNA molecule. However, even if its surface represents a site for the RNA–protein contact, β-sheet plays no essential role in the discrimination between RNA sequences, as this is the function of regions formed by the closely located hydrophobic amino acid side chains that position RNA bases. Positively charged and polar side chains entering these pockets form ionic and hydrogen bonds with the atoms of nitrogenous bases, thereby ensuring specificity of nucleotide recognition. The pockets are often formed by additional structural elements that do not belong to the conserved protein motifs. Specific pockets can be also formed in the absence of β-sheet, although the principles of RNA base positioning via stacking with aromatic residues and recognition due to the contacts with polar/charged side chains are preserved in this case.
Studying the spatial structures of proteins in complex with convoluted spatial structures of RNA revealed that proteins have a small number of contacts with the RNA bases and that almost all contacts are formed by the atoms of the RNA sugar-phosphate backbone. This can be explained by the conformational features of the A-form of dsRNA, which has a deep and closed from external contacts major groove that impedes direct contact of the protein residues with the dsRNA bases. At the same time, the minor groove is shallow and wide. This facilitates access to the nucleotides, but simultaneously increases accessibility of the potential RNA–protein contacts to the external factors (water molecules and ions) and prevents energy gain during the complex formation. As a result, the contacts between the protein atoms and atoms of the sugar-phosphate backbone dominate, while recognition of the nucleotide sequences is hindered or even impossible. Instead, spatial RNA structures are recognized, which is the most pronounced difference in the interactions of RNA-binding proteins with ssRNAs and dsRNAs
Another feature of the ssRNA-recognizing proteins is binding of short (4 to 6 nt) RNA sequences. Such short region of contact is sometimes insufficient for the tight RNA binding. Hence, the ssRNA-binding proteins often contain repeated (duplicated) RNA-binding domains, which allows to significantly expand the region of contact, to increase stability of the formed RNA–protein complexes, and to ensure recognition of a larger number of different sequences.
dsRNA-binding proteins recognize unique stable RNA spatial structures. These proteins contain preformed complementary areas on their surface [116]. The requirement for complementarity of the RNA–protein contacts results in the unique nature of the dsRNA-binding structures in these proteins, except proteins with the dsRBD domain proteins that interact with dsRNAs containing single bulging nucleotides. All other known dsDNA-binding proteins recognize unique RNA spatial structures, which provides their selectivity and affinity toward interacting molecules.
Acknowledgments. The author is grateful to S. V. Tishchenko for helpful discussion during writing of this review and to S. V. Nikonov, M. B. Garber, and N. A. Nevskaya for the fundamental studies that served as a foundation for this work.
Ethics declarations. The author declares no conflict of interest in financial or any other sphere. The article contains no description of studies involving animals or humans performed by the author.
REFERENCES
1.Fedorov, R., Meshcheryakov, V., Gongadze, G.,
Fomenkova, N., Nevskaya, N., et al. (2001) Structure of ribosomal
protein TL5 complexed with RNA provides new insights into the CTC
family of stress proteins, Acta Crystallogr. Sect. D Biol.
Crystallogr., 57, 968-976, doi:
10.1107/S0907444901006291.
2.Perederina, A., Nevskaya, N., Nikonov, O., Nikulin,
A., Dumas, P., et al. (2002) Detailed analysis of RNA–protein
interactions within the bacterial ribosomal protein L5/5S rRNA complex,
RNA, 8, 1548-1557, doi: 10.1017/s1355838202029953.
3.Nikulin, A., Eliseikina, I., Tishchenko, S.,
Nevskaya, N., Davydova, N., et al. (2003) Structure of the L1
protuberance in the ribosome, Nat. Struct. Biol., 10,
104-108, doi: 10.1038/nsb886.
4.Tishchenko, S., Nikulin, A., Fomenkova, N.,
Nevskaya, N., Nikonov, O., et al. (2001) Detailed analysis of
RNA–protein interactions within the ribosomal protein
S8–rRNA complex from the Archaeon Methanococcus
jannaschii, J. Mol. Biol., 311, 311-324, doi:
10.1006/jmbi.2001.4877.
5.Nikulin, A., Serganov, A., Ennifar, E., Tishchenko,
S., Nevskaya, N., et al. (2000) Crystal structure of the S15–rRNA
complex, Nat. Struct. Biol., 7, 273-277, doi:
10.1038/74028.
6.Gabdulkhakov, A., Mitroshin, I., and Garber, M.
(2020) Structure of the ribosomal P stalk base in archaean
Methanococcus jannaschii, J. Struct. Biol.,
211, 107559, doi: 10.1016/j.jsb.2020.107559.
7.Kostareva, O. S., Nevskaya, N. A., Tishchenko, S.
V., Gabdulkhakov, A. G., Garber, M. B., and Nikonov, S. V. (2018)
Influence of nonconserved regions of L1 protuberance of Thermus
thermophilus ribosome on the affinity of L1 protein to 23s rRNA,
Mol. Biol., 52, 91-95, doi:
10.1134/S0026893318010090.
8.Draper, D. E. (1999) Themes in RNA–protein
recognition, J. Mol. Biol., 293, 255-270, doi:
10.1006/jmbi.1999.2991.
9.Corley, M., Burns, M. C., and Yeo, G. W. (2020) How
RNA-binding proteins interact with RNA: molecules and mechanisms,
Mol. Cell, 78, 9-29, doi:
10.1016/j.molcel.2020.03.011.
10.Iacobuzio-Donahue, C. A., Ashfaq, R., Maitra, A.,
Adsay, N. V., Shen-Ong, G. L., et al. (2003) Highly expressed genes in
pancreatic ductal adenocarcinomas: a comprehensive characterization and
comparison of the transcription profiles obtained from three major
technologies, Cancer Res., 63, 8614-8622, doi:
10.1126/science.1058040.
11.Swanson, M. S., Nakagawa, T. Y., LeVan, K., and
Dreyfuss, G. (1987) Primary structure of human nuclear
ribonucleoprotein particle C proteins: conservation of sequence and
domain structures in heterogeneous nuclear RNA, mRNA, and
pre-rRNA-binding proteins, Mol. Cell. Biol., 7,
1731-1739, doi: 10.1128/MCB.7.5.1731.
12.Sillekens, P. T. G., Habets, W. J., Beijer, R.
P., and van Venrooij, W. J. (1987) CDNA cloning of the human U1
SnRNA-associated A protein: extensive homology between U1 and U2
SnRNP-specific proteins, EMBO J., 6, 3841-3848.
13.Muto, Y., and Yokoyama, S. (2012) Structural
insight into RNA recognition motifs: versatile molecular lego building
blocks for biological systems, Wiley Interdisc. Rev. RNA,
3, 229-246, doi: 10.1002/wrna.1107.
14.Messias, A. C., and Sattler, M. (2004) Structural
basis of single-stranded RNA recognition, Accounts Chem. Res.,
37, 279-287, doi: 10.1021/ar030034m.
15.Cléry, A., Blatter, M., and Allain, F.
H.-T. (2008) RNA recognition motifs: boring? Not quite, Curr. Opin.
Struct. Biol., 18, 290-298, doi:
10.1016/j.sbi.2008.04.002.
16.León, B., Kashyap, M. K., Chan, W. C.,
Krug, K. A., Castro, J. E., et al. (2017) A challenging pie to splice:
drugging the spliceosome, Angewandte Chemie Int. Edn.,
56, 12052-12063, doi: 10.1002/anie.201701065.
17.Van der Feltz, C., Anthony, K., Brilot, A., and
Pomeranz Krummel, D. A. (2012) Architecture of the spliceosome,
Biochemistry, 51, 3321-3333, doi: 10.1021/bi201215r.
18.Mazza, C., Segref, A., Mattaj, I. W., and Cusack,
S. (2002) Large-scale induced fit recognition of an m(7)GpppG Cap
analogue by the human nuclear Cap-binding complex, EMBO J.,
21, 5548-5557, doi: 10.1093/emboj/cdf538.
19.Calero, G., Wilson, K. F., Ly, T., Rios-Steiner,
J. L., Clardy, J. C., and Cerione, R. A. (2002) Structural basis of
M7GpppG Binding to the nuclear Cap-binding protein complex, Nat.
Struct. Biol., 9, 912-917, doi: 10.1038/nsb874.
20.Johansson, C., Finger, L. D., Trantirek, L.,
Mueller, T. D., Kim, S., et al. (2004) Solution structure of the
complex formed by the two N-terminal RNA-binding domains of nucleolin
and a pre-rRNA target, J. Mol. Biol., 337, 799-816, doi:
10.1016/j.jmb.2004.01.056.
21.Allain, F. H., Bouvet, P., Dieckmann, T., and
Feigon, J. (2000) Molecular basis of sequence-specific recognition of
pre-ribosomal RNA by nucleolin, EMBO J., 19, 6870-6881,
doi: 10.1093/emboj/19.24.6870.
22.Price, S. R., Evans, P. R., and Nagai, K. (1998)
Crystal structure of the spliceosomal
U2B′′–U2A′ protein complex bound to a fragment
of U2 small nuclear RNA, Nature, 394, 645-650, doi:
10.1038/29234.
23.Oubridge, C., Ito, N., Evans, P. R., Teo, C. H.,
and Nagai, K. (1994) Crystal structure at 1.92 Å resolution of
the RNA-binding domain of the U1A spliceosomal protein complexed with
an RNA hairpin, Nature, 372, 432-438, doi:
10.1038/372432a0.
24.Handa, N., Nureki, O., Kurimoto, K., Kim, I.,
Sakamoto, H., et al. (1999) Structural basis for recognition of the Tra
mRNA precursor by the sex-lethal protein, Nature, 398,
579-584, doi: 10.1038/19242.
25.Afroz, T., Cienikova, Z., Cléry, A., and
Allain, F. H. T. (2015) One, two, three, four! How multiple RRMs read
the genome sequence, Methods Enzymol., 558,
235-278, doi: 10.1016/bs.mie.2015.01.015.
26.Grishin, N. V. (2001) KH domain: one motif, two
folds, Nucleic Acids Res., 29, 638-643, doi:
10.1093/nar/29.3.638.
27.Oddone, A., Lorentzen, E., Basquin, J., Gasch,
A., Rybin, V., et al. (2007) Structural and biochemical
characterization of the yeast exosome component Rrp40, EMBO
Rep., 8, 63-69, doi: 10.1038/sj.embor.7400856.
28.Auweter, S. D., Oberstrass, F. C., and Allain, F.
H.-T. (2006) Sequence-specific binding of single-stranded RNA: is there
a code for recognition? Nucleic Acids Res., 34,
4943-4959, doi: 10.1093/nar/gkl620.
29.Nicastro, G., Taylor, I. A., and Ramos, A. (2015)
KH-RNA interactions: back in the groove, Curr. Opin. Struct.
Biol., 30, 63-70, doi: 10.1016/j.sbi.2015.01.002.
30.Lewis, H. A., Musunuru, K., Jensen, K. B., Edo,
C., Chen, H., et al. (2000) Sequence-specific RNA binding by a nova KH
domain, Cell, 100, 323-332, doi:
10.1016/S0092-8674(00)80668-6.
31.Ryder, S. P., and Massi, F. (2010) Insights into
the structural basis of RNA recognition by star domain proteins,
Adv. Exp. Med. Biol., 693, 37-53, doi:
10.1007/978-1-4419-7005-3_3.
32.Beuth, B., Pennell, S., Arnvig, K. B., Martin, S.
R., and Taylor, I. A. (2005) Structure of a Mycobacterium
tuberculosis NusA–RNA complex, EMBO J., 24,
3576-3587, doi: 10.1038/sj.emboj.7600829.
33.Lunde, B. M., Moore, C., and Varani, G. (2007)
RNA-binding proteins: modular design for efficient function, Nat.
Rev. Mol. Cell Biol., 8, 479-490, doi: 10.1038/nrm2178.
34.Valverde, R., Edwards, L., and Regan, L. (2008)
Structure and function of KH domains, FEBS J., 275,
2712-2726, doi: 10.1111/j.1742-4658.2008.06411.x.
35.Diakun, G. P., Fairall, L., and Klug, A. (1986)
EXAFS study of the zinc-binding sites in the protein transcription
factor IIIA, Nature, 324, 698-699, doi:
10.1038/324698a0.
36.Ginsberg, A. M., King, B. O., and Roeder, R. G.
(1984) Xenopus 5S gene transcription factor, TFIIIA: characterization
of a CDNA clone and measurement of RNA levels throughout development,
Cell, 39, 479-489, doi: 10.1016/0092-8674(84)90455-0.
37.Miller, J., McLachlan, A. D., and Klug, A. (1985)
Repetitive zinc-binding domains in the protein transcription factor
IIIA from Xenopus oocytes, EMBO J., 4,
1609-1614.
38.Hudson, B. P., Martinez-Yamout, M. A., Dyson, H.
J., and Wright, P. E. (2004) Recognition of the mRNA au-rich element by
the zinc finger domain of TIS11d, Nat. Struct. Mol. Biol.,
11, 257-264, doi: 10.1038/nsmb738.
39.Iwanaga, E., Nanri, T., Mitsuya, H., and Asou, N.
(2011) Mutation in the RNA binding protein TIS11D/ZFP36L2 is associated
with the pathogenesis of acute leukemia, Int. J. Oncology,
38, 25-31, doi: 10.3892/ijo_00000820.
40.Dey, A., York, D., Smalls-Mantey, A., and
Summers, M. F. (2005) Composition and sequence-dependent binding of RNA
to the nucleocapsid protein of Moloney murine leukemia virus,
Biochemistry, 44, 3735-3744, doi: 10.1021/bi047639q.
41.D’Souza, V., and Summers, M. F. (2004)
Structural basis for packaging the dimeric genome of Moloney murine
leukaemia virus, Nature, 431, 586-590, doi:
10.1038/nature02944.
42.Wang, X., McLachlan, J., Zamore, P. D., and Hall,
T. M. T. (2002) Modular recognition of RNA by a human pumilio-homology
domain, Cell, 110, 501-512, doi:
10.1016/s0092-8674(02)00873-5.
43.De Moor, C. H., Meijer, H., and Lissenden, S.
(2005) Mechanisms of translational control by the 3′-UTR in
development and differentiation, Semin. Cell Dev. Biol.,
16, 49-58, doi: 10.1016/j.semcdb.2004.11.007.
44.Wickens, M., Bernstein, D. S., Kimble, J., and
Parker, R. (2002) A PUF family portrait: 3′UTR regulation as a
way of life, Trends Genet. TIG, 18, 150-157, doi:
10.1016/s0168-9525(01)02616-6.
45.Mackay, J. P., Font, J., and Segal, D. J. (2011)
The prospects for designer single-stranded RNA-binding proteins,
Nat. Struct. Mol. Biol., 18, 256-261, doi:
10.1038/nsmb.2005.
46.Cheong, C.-G., and Hall, T. M. T. (2006)
Engineering RNA sequence specificity of pumilio repeats, Proc. Natl.
Acad. Sci. USA, 103, 13635-13639, doi:
10.1073/pnas.0606294103.
47.Antson, A. A., Dodson, E. J., Dodson, G.,
Greaves, R. B., Chen, X., and Gollnick, P. (1999) Structure of the Trp
RNA-binding attenuation protein, TRAP, bound to RNA, Nature,
401, 235-242, doi: 10.1038/45730.
48.Babitzke, P. (1997) Regulation of tryptophan
biosynthesis: Trp-Ing the TRAP or how Bacillus subtilis
reinvented the wheel, Mol. Microbiol., 26, 1-9, doi:
10.1046/j.1365-2958.1997.5541915.x.
49.Elliott, M. B., Gottlieb, P. A., and Gollnick, P.
(1999) Probing the TRAP-RNA interaction with nucleoside analogs,
RNA, 5, 1277-1289, doi: 10.1017/s1355838299991057.
50.Theobald, D. L., Mitton-Fry, R. M., and Wuttke,
D. S. (2003) Nucleic acid recognition by OB-fold proteins, Annu.
Rev. Biophys. Biomol. Struct., 32, 115-133, doi:
10.1146/annurev.biophys.32.110601.142506.
51.Skordalakes, E., and Berger, J. M. (2003)
Structure of the Rho transcription terminator, Cell, 114,
135-146, doi: 10.1016/S0092-8674(03)00512-9.
52.Bogden, C. E., Fass, D., Bergman, N., Nichols, M.
D., and Berger, J. M. (1999) The structural basis for terminator
recognition by the Rho transcription termination factor, Mol.
Cell, 3, 487-493, doi: 10.1016/S1097-2765(00)80476-1.
53.Keto-Timonen, R., Hietala, N., Palonen, E.,
Hakakorpi, A., Lindström, M., and Korkeala, H. (2016) Cold shock
proteins: a minireview with special emphasis on Csp-family of
enteropathogenic yersinia, Front. Microbiol., 7, doi:
10.3389/fmicb.2016.01151.
54.Horn, G., Hofweber, R., Kremer, W., and
Kalbitzer, H. R. (2007) Structure and function of bacterial cold shock
proteins, Cell. Mol. Life Sci., 64, 1457-1470, doi:
10.1007/s00018-007-6388-4.
55.Amir, M., Kumar, V., Dohare, R., Islam, A.,
Ahmad, F., and Hassan, M. I. (2018) Sequence, structure and
evolutionary analysis of cold shock domain proteins, a member of OB
Fold family, J. Evol. Biol., 31, 1903-1917, doi:
10.1111/jeb.13382.
56.Lyabin, D. N., Eliseeva, I. A., and Ovchinnikov,
L. P. (2014) YB-1 protein: functions and regulation, RNA,
5, 95-110, doi: 10.1002/wrna.1200.
57.Hermann, H., Fabrizio, P., Raker, V. A., Foulaki,
K., Hornig, H., et al. (1995) SnRNP Sm proteins share two
evolutionarily conserved sequence motifs which are involved in sm
protein–protein interactions, EMBO J., 14,
2076-2088.
58.Pomeranz Krummel, D. A., Oubridge, C., Leung, A.
K. W., Li, J., and Nagai, K. (2009) Crystal structure of human
spliceosomal U1 SnRNP at 5.5 Å resolution, Nature,
458, 475-480, doi: 10.1038/nature07851.
59.Yong, J., Golembe, T. J., Battle, D. J.,
Pellizzoni, L., and Dreyfuss, G. (2004) SnRNAs contain specific
SMN-binding domains that are essential for SnRNP assembly, Mol.
Cell. Biol., 24, 2747-2756, doi:
10.1128/MCB.24.7.2747-2756.2004.
60.Kondo, Y., Oubridge, C., van Roon, A.-M. M., and
Nagai, K. (2015) Crystal structure of human U1 SnRNP, a small nuclear
ribonucleoprotein particle, reveals the mechanism of 5′ splice
site recognition, eLife, 4, doi: 10.7554/eLife.04986.
61.Mura, C., Randolph, P. S., Patterson, J., and
Cozen, A. E. (2013) Archaeal and eukaryotic homologs of Hfq, RNA
Biol., 10, 636-651, doi: 10.4161/rna.24538.
62.He, W., and Parker, R. (2000) Functions of Lsm
proteins in mRNA degradation and splicing, Curr. Opin. Cell
Biol., 12, 346-350, doi: 10.1016/S0955-0674(00)00098-3.
63.Sharif, H., and Conti, E. (2013) Architecture of
the Lsm1-7-Pat1 complex: a conserved assembly in eukaryotic mRNA
turnover, Cell Rep., 5, 283-291, doi:
10.1016/j.celrep.2013.10.004.
64.Sauer, E. (2013) Structure and RNA-binding
properties of the bacterial LSm protein Hfq, RNA Biol.,
10, 610-618, doi: 10.4161/rna.24201.
65.Murina, V. N., and Nikulin, A. D. (2011)
RNA-binding Sm-electrical proteins of bacteria and archaea: similarity
and structure of structures and functions [in Russian], Usp. Biol.
Khim., 51, 133-164.
66.Schumacher, M. A., Pearson, R. F., Møller,
T., Valentin-Hansen, P., and Brennan, R. G. (2002) Structures of the
pleiotropic translational regulator Hfq and an Hfq–RNA complex: a
bacterial sm-like protein, EMBO J., 21, 3546-3556, doi:
10.1093/emboj/cdf322.
67.Link, T. M., Valentin-Hansen, P., and Brennan, R.
G. (2009) Structure of Escherichia coli Hfq bound to
polyriboadenylate RNA, Proc. Natl. Acad. Sci. USA, 106,
19292-19297, doi: 10.1073/pnas.0908744106.
68.Vogel, J., and Luisi, B. F. (2011) Hfq and its
constellation of RNA, Nat. Rev. Microbiol., 9, 578-589,
doi: 10.1038/nrmicro2615.
69.Someya, T., Baba, S., Fujimoto, M., Kawai, G.,
Kumasaka, T., and Nakamura, K. (2012) Crystal structure of Hfq from
Bacillus subtilis in complex with SELEX-derived RNA
aptamer: insight into RNA-binding properties of bacterial Hfq,
Nucleic Acids Res., 40, 1856-1867, doi:
10.1093/nar/gkr892.
70.Masliah, G., Barraud, P., and Allain, F. H.-T.
(2012) RNA recognition by double-stranded RNA binding domains: a matter
of shape and sequence, Cell. Mol. Life Sci., 70,
1875-1895, doi: 10.1007/s00018-012-1119-x.
71.Gleghorn, M. L., and Maquat, L. E. (2014)
“Black sheep” that don’t leave the double-stranded
RNA-binding domain fold, Trends Biochem. Sci., 39,
328-340, doi: 10.1016/j.tibs.2014.05.003.
72.St. Johnston, D., Brown, N. H., Gall, J. G., and
Jantsch, M. (1992) A conserved double-stranded RNA-binding domain,
Proc. Natl. Acad. Sci. USA, 89, 10979-10983, doi:
10.1073/pnas.89.22.10979.
73.Ryter, J. M., and Schultz, S. C. (1998) Molecular
basis of double-stranded RNA–protein interactions: structure of a
DsRNA-binding domain complexed with DsRNA, EMBO J., 17,
7505-7513, doi: 10.1093/emboj/17.24.7505.
74.Stefl, R., Xu, M., Skrisovska, L., Emeson, R. B.,
and Allain, F. H.-T. (2006) Structure and specific RNA binding of ADAR2
double-stranded RNA binding motifs, Structure, 14,
345-355, doi: 10.1016/j.str.2005.11.013.
75.Spillmann, S., Dohme, F., and Nierhaus, K. H.
(1977) Assembly in vitro of the 50 S subunit from
Escherichia coli ribosomes: proteins essential for the
first heat-dependent conformational change, J. Mol. Biol.,
115, 513-523, doi: 10.1016/0022-2836(77)90168-1.
76.Held, W. A., and Nomura, M. (1973) Structure and
function of bacterial ribosomes. XX. Rate-determining step in the
reconstitution of Escherichia coli 30S ribosomal
subunits, Biochemistry, 12, 3273-3281, doi:
10.1021/bi00741a020.
77.Nikulin, A. D. (2002) Studies of interactions of
ribosomal proteins with ribosomal RNAs, Usp. Biol. Khim.,
42, 61-88.
78.Nikulin, A. D. (2018) Structural aspects of
ribosomal RNA recognition by ribosomal proteins, Biochemistry
(Moscow), 83, S111-S133, doi: 10.1134/S0006297918140109.
79.Ban, N., Beckmann, R., Cate, J. H., Dinman, J.
D., Dragon, F., et al. (2014) A new system for naming ribosomal
proteins, Curr. Opin. Struct. Biol., 24, 165-169, doi:
10.1016/j.sbi.2014.01.002.
80.Held, W. A., Ballou, B., Mizushima, S., and
Nomura, M. (1974) Assembly mapping of 30 S ribosomal proteins from
Escherichia coli. Further studies, J. Biol. Chem.,
249, 3103-3111.
81.Gregory, R. J., Zeller, M. L., Thurlow, D. L.,
Gourse, R. L., Stark, M. J. R., et al. (1984) Interaction of ribosomal
proteins S6, S8, S15 and S18 with the central domain of 16 S ribosomal
RNA from Escherichia coli, J. Mol. Biol.,
178, 287-302, doi: 10.1016/0022-2836(84)90145-1.
82.Mougel, M., Eyermann, F., Westhof, E., Romby, P.,
Expert-Bezançon, A., et al. (1987) Binding of Escherichia
coli ribosomal protein S8 to 16 S rRNA. A model for the
interaction and the tertiary structure of the RNA binding site, J.
Mol. Biol., 198, 91-107, doi:
10.1016/0022-2836(87)90460-8.
83.Svensson, P., Changchien, L. M., Craven, G. R.,
and Noller, H. F. (1988) Interaction of ribosomal proteins, S6, S8, S15
and S18 with the central domain of 16 S ribosomal RNA, J. Mol.
Biol., 200, 301-308, doi: 10.1016/0022-2836(88)90242-2.
84.Chen, S. S., Sperling, E., Silverman, J. M.,
Davis, J. H., and Williamson, J. R. (2012) Measuring the dynamics of
E. coli ribosome biogenesis using pulse-labeling and
quantitative mass spectrometry, Mol. bioSystems, 8,
3325-3334, doi: 10.1039/c2mb25310k.
85.Menichelli, E., Edgcomb, S. P., Recht, M. I., and
Williamson, J. R. (2012) The structure of aquifex aeolicus ribosomal
protein S8 reveals a unique subdomain that contributes to an extremely
tight association with 16S rRNA, J. Mol. Biol., 415,
489-502, doi: 10.1016/j.jmb.2011.10.046.
86.Geyl, D., Böck, A., and Wittmann, H. G.
(1977) Cold-sensitive growth of a mutants of Escherichia
coli with an altered ribosomal protein S8: analysis of
revertants, Mol. Gen. Genet., 152, 331-336, doi:
10.1007/BF00693088.
87.Powers, T., and Noller, H. F. (1995) Hydroxyl
radical footprinting of ribosomal proteins on 16S rRNA, RNA,
1, 194-209.
88.Moine, H., Cachia, C., Westhof, E., Ehresmann,
B., and Ehresmann, C. (1997) The RNA binding site of S8 ribosomal
protein of Escherichia coli: selex and hydroxyl radical
probing studies, RNA, 3, 255-268.
89.Kalurachchi, K., Uma, K., Zimmermann, R. A., and
Nikonowicz, E. P. (1997) Structural features of the binding site for
ribosomal protein S8 in Escherichia coli 16S rRNA defined
using NMR spectroscopy, Proc. Natl. Acad. Sci. USA, 94,
2139-2144, doi: 10.1073/pnas.94.6.2139.
90.Gregory, R. J., Cahill, P. B. F., Thurlow, D. L.,
and Zimmermann, R. A. (1988) Interaction of Escherichia
coli ribosomal protein S8 with its binding sites in ribosomal
RNA and messenger RNA, J. Mol. Biol., 204, 295-307, doi:
10.1016/0022-2836(88)90577-3.
91.Yates, J. L., Arfsten, A. E., and Nomura, M.
(1980) In vitro expression of Escherichia coli
ribosomal protein genes: autogenous inhibition of translation, Proc.
Natl. Acad. Sci. USA, 77, 1837-1841, doi:
10.1073/pnas.77.4.1837.
92.Cerretti, D. P., Mattheakis, L. C., Kearney, K.
R., Vu, L., and Nomura, M. (1988) Translational regulation of the spc
operon in Escherichia coli, J. Mol. Biol.,
204, 309-325, doi: 10.1016/0022-2836(88)90578-5.
93.Olins, P. O., and Nomura, M. (1981) Translational
regulation by ribosomal protein S8 in Escherichia coli:
structural homology between rRNA binding site and feedback target on
mRNA, Nucleic Acids Res., 9, 1757-1764, doi:
10.1093/nar/9.7.1757.
94.Merianos, H. J., Wang, J., and Moore, P. B.
(2004) The structure of a ribosomal protein S8/Spc operon mRNA complex,
RNA, 10, 954-964, doi: 10.1261/rna.7030704.
95.Agalarov, S. C., Sridhar Prasad, G., Funke, P.
M., Stout, C. D., and Williamson, J. R. (2000) Structure of the
S15,S6,S18-rRNA complex: assembly of the 30S ribosome central domain,
Science, 288, 107-113, doi:
10.1126/science.288.5463.107.
96.Duss, O., Stepanyuk, G. A., Grot, A.,
O’Leary, S. E., Puglisi, J. D., and Williamson, J. R. (2018)
Real-time assembly of ribonucleoprotein complexes on nascent RNA
transcripts, Nat. Commun., 9, 5087, doi:
10.1038/s41467-018-07423-3.
97.Portier, C., Dondon, L., and Grunberg-Manago, M.
(1990) Translational autocontrol of the Escherichia coli
ribosomal protein S15, J. Mol. Biol., 211, 407-414, doi:
10.1016/0022-2836(90)90361-O.
98.Portier, C., Philippe, C., Dondon, L.,
Grunberg-Manago, M., Ebel, J. P., et al. (1990) Translational control
of ribosomal protein S15, Biochim. Biophys. Acta Gene Struct.
Express., 1050, 328-336, doi:
10.1016/0167-4781(90)90190-D.
99.Philippe, C., Eyermann, F., Benard, L., Portier,
C., Ehresmann, B., and Ehresmann, C. (1993) Ribosomal protein S15 from
Escherichia coli modulates its Own translation by
trapping the ribosome on the mRNA initiation loading site, Proc.
Natl. Acad. Sci. USA, 90, 4394-4398, doi:
10.1073/pnas.90.10.4394.
100.Philippe, C., Bénard, L., Eyermann, F.,
Cachia, C., Kirillov, S. V., et al. (1994) Structural elements of RpsO
mRNA involved in the modulation of translational initiation and
regulation of E. coli ribosomal protein S15, Nucleic Acids
Res., 22, 2538-2546, doi: 10.1093/nar/22.13.2538.
101.Mougel, M., Philippe, C., Ebel, J. P.,
Ehresmann, B., and Ehresmann, C. (1988) The E. coli 16S rRNA
binding site of ribosomal protein S15: higher-order structure in the
absence and in the presence of the protein, Nucleic Acids Res.,
16, 2825-2839.
102.Serganov, A., Ennifar, E., Portier, C.,
Ehresmann, B., and Ehresmann, C. (2002) Do mRNA and rRNA binding sites
of E. coli ribosomal protein S15 share common structural
determinants? J. Mol. Biol., 320, 963-978, doi:
10.1016/S0022-2836(02)00553-3.
103.Serganov, A., Polonskaia, A., Ehresmann, B.,
Ehresmann, C., and Patel, D. J. (2003) Ribosomal protein S15 represses
its own translation via adaptation of an rRNA-like fold within its
mRNA, EMBO J., 22, 1898-1908, doi:
10.1093/emboj/cdg170.
104.Springer, M., and Portier, C. (2003) More than
one way to skin a cat: translational autoregulation by ribosomal
protein S15, Nat. Struct. Biol., 10, 420-422, doi:
10.1038/nsb0603-420.
105.Mathy, N., Pellegrini, O., Serganov, A., Patel,
D. J., Ehresmann, C., and Portier, C. (2004) Specific recognition of
RpsO mRNA and 16S rRNA by Escherichia coli ribosomal
protein S15 relies on both mimicry and site differentiation, Mol.
Microbiol., 52, 661-675, doi:
10.1111/j.1365-2958.2004.04005.x.
106.Ehresmann, C., Ehresmann, B., Ennifar, E.,
Dumas, P., Garber, M., et al. (2004) Molecular mimicry in translational
regulation: the case of ribosomal protein S15, RNA Biol.,
1, 66-73, doi: 10.4161/rna.1.1.958.
107.Nikonov, S., Nevskaya, N., Eliseikina, I.,
Fomenkova, N., Nikulin, A., et al. (1996) Crystal structure of the RNA
binding ribosomal protein L1 from Thermus thermophilus,
EMBO J., 15, 1350-1359.
108.Brot, N., Caldwell, P., and Weissbach, H.
(1981) Regulation of synthesis of Escherichia coli
ribosomal proteins L1 and L11, Arch. Biochem. Biophys.,
206, 51-53, doi: 10.1016/0003-9861(81)90064-3.
109.Yates, J. (1981) Feedback regulation of
ribosomal protein synthesis in E. coli: localization of the mRNA
target sites for repressor action of ribosomal protein L1, Cell,
24, 243-249, doi: 10.1016/0092-8674(81)90520-1.
110.Tishchenko, S., Nikonova, E., Kostareva, O.,
Gabdulkhakov, A., Piendl, W., et al. (2011) Structural analysis of
interdomain mobility in ribosomal L1 proteins, Acta Crystallograp.
Sect. D, 67, 1023-1027, doi: 10.1107/S0907444911043435.
111.Nevskaya, N. (2005) Ribosomal protein L1
recognizes the same specific structural motif in its target sites on
the autoregulatory mRNA and 23S rRNA, Nucleic Acids Res.,
33, 478-485, doi: 10.1093/nar/gki194.
112.Nevskaya, N., Tishchenko, S., Volchkov, S.,
Kljashtorny, V., Nikonova, E., et al. (2006) New insights into the
interaction of ribosomal protein L1 with RNA, J. Mol. Biol.,
355, 747-759, doi: 10.1016/j.jmb.2005.10.084.
113.Tishchenko, S., Nikonova, E., Nikulin, A.,
Nevskaya, N., Volchkov, S., et al. (2006) Structure of the ribosomal
protein L1–mRNA complex at 2.1 Å resolution: common
features of crystal packing of L1-RNA complexes, Acta Crystallogr.
Sect. D Biol. Crystallogr., 62, 1545-1554, doi:
10.1107/S0907444906041655.
114.Tishchenko, S., Nikonova, E., Kljashtorny, V.,
Kostareva, O., Nevskaya, N., et al. (2007) Domain I of ribosomal
protein L1 is sufficient for specific RNA binding, Nucleic Acids
Res., 35, 7389-95, doi: 10.1093/nar/gkm898.
115.Tishchenko, S., Kostareva, O., Gabdulkhakov,
A., Mikhaylina, A., Nikonova, E., et al. (2015) Protein–RNA
affinity of ribosomal protein L1 mutants does not correlate with the
number of intermolecular interactions, Acta Crystallogr. Sect. D
Biol. Crystallogr., 71, 376-386, doi:
10.1107/S1399004714026248.
116.Nevskaya, N. A., Nikonov, O. S., Revtovich, S.
V., Garber, M. B., and Nikonov, S. V. (2004) Identification of
RNA-recognizing modules on the surface of ribosomal proteins, Mol.
Biol., 38, 789-798, doi:
10.1023/B:MBIL.0000043948.74962.05.